12.3 Bacterial Replication Requires a Large Number of Enzymes and Proteins
Replication takes place in four stages: initiation, unwinding, elongation, and termination. The following discussion of the process of replication will focus on bacterial systems, where replication has been most thoroughly studied and is best understood. Although many aspects of replication in eukaryotic cells are similar to those in bacterial cells, there are some important differences. We will compare bacterial and eukaryotic replication later in this chapter.
Initiation
The circular chromosome of E. coli has a single replication origin (oriC). The minimal sequence required for oriC to function consists of 245 bp that contain several critical sites. An initiator protein (known as DnaA in E. coli) binds to oriC and causes a short section of DNA to unwind. This unwinding allows helicase and other single-strand-binding proteins to attach to the polynucleotide strand (Figure 12.11).
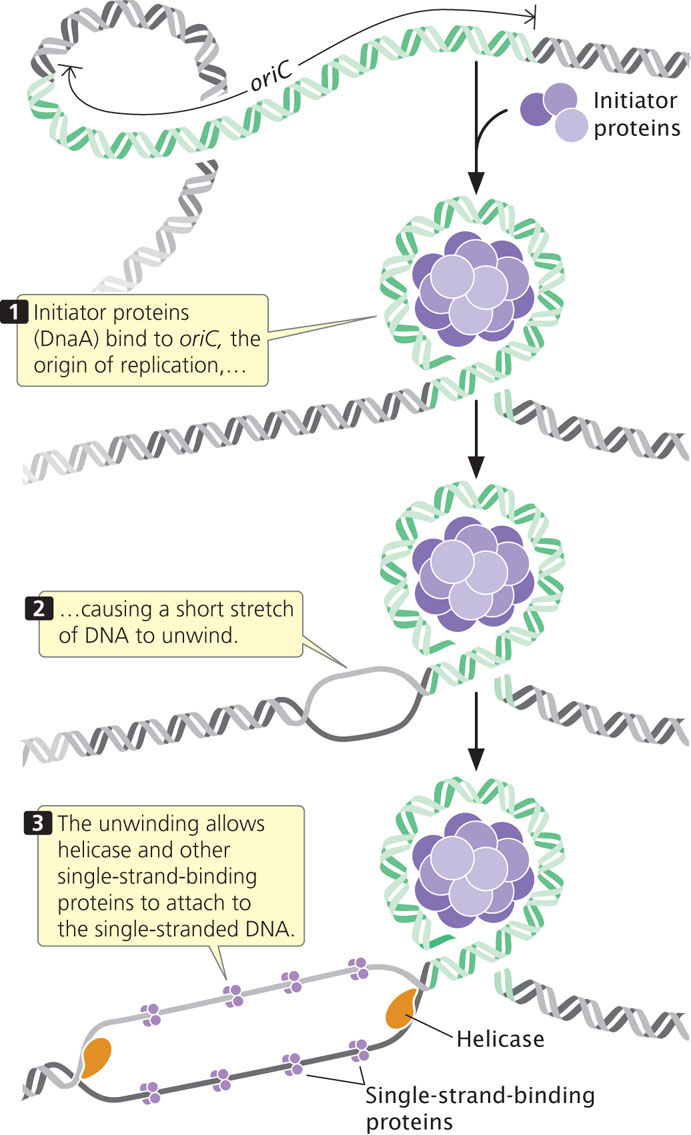
Unwinding
Because DNA synthesis requires a single-stranded template and because double-stranded DNA must be unwound before DNA synthesis can take place, the cell relies on several proteins and enzymes to accomplish the unwinding.
DNA Helicase
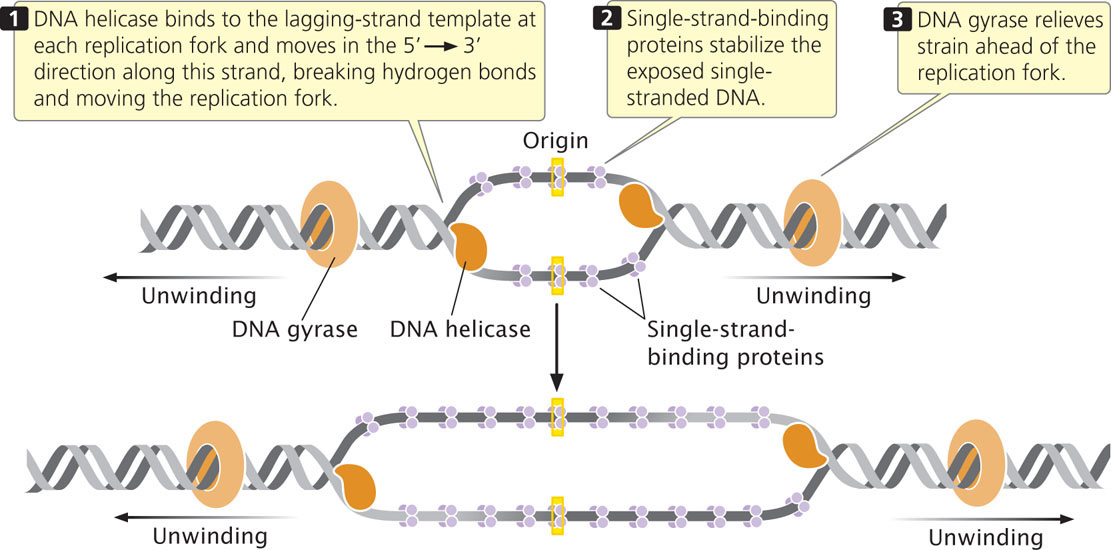
A DNA helicase breaks the hydrogen bonds that exist between the bases of the two nucleotide strands of a DNA molecule. Helicase cannot initiate the unwinding of double-stranded DNA; the initiator protein first separates DNA strands at the origin, providing a short stretch of single-stranded DNA to which a helicase binds. Helicase binds to the lagging-strand template at each replication fork and moves in the 5′→3′ direction along this strand, thus also moving the replication fork (Figure 12.12).
Single-Strand-Binding Proteins
After DNA has been unwound by helicase, single-strand-binding proteins (SSBs) attach tightly to the exposed single-stranded DNA (see Figure 12.12). These proteins protect the single-stranded nucleotide chains and prevent the formation of secondary structures such as hairpins (see Figure 10.17) that interfere with replication. Unlike many DNA-binding proteins, SSBs are indifferent to base sequence: they will bind to any single-stranded DNA. Single-strand-binding proteins form tetramers (groups of four); each tetramer covers from 35 to 65 nucleotides.
DNA Gyrase
Another protein essential for the unwinding process is the enzyme DNA gyrase, a topoisomerase. As discussed in Chapter 11 and the introduction to this chapter, topoisomerases control the supercoiling of DNA. They come in two major types: type I topoisomerases alter supercoiling by making single-strand breaks in DNA, while type II topoisomerases create double-stranded breaks. DNA gyrase is a type II topiosmerase. In replication, it reduces the torsional strain (torque) that builds up ahead of the replication fork as a result of unwinding (see Figure 12.12). It reduces torque by making a double-stranded break in one segment of the DNA helix, passing another segment of the helix through the break, and then resealing the broken ends of the DNA. This action, which requires ATP, removes a twist in the DNA and reduces the supercoiling.
A group of antibiotics called 4-quinolones kill bacteria by binding to DNA gyrase and inhibiting its action. The inhibition of DNA gyrase results in the cessation of DNA synthesis and bacterial growth. An example of a 4-quinolone is nalidixic acid, which was first introduced in the 1960s and is commonly used to treat urinary infections. Many bacteria have acquired resistance to quinolones through mutations in the gene for DNA gyrase.
CONCEPTS
Replication is initiated at a replication origin, where an initiator protein binds and causes a short stretch of DNA to unwind. DNA helicase breaks hydrogen bonds at a replication fork, and single-strand-binding proteins stabilize the separated strands. DNA gyrase reduces the torsional strain that develops as the two strands of double-helical DNA unwind.
 CONCEPT CHECK 4
CONCEPT CHECK 4Place the following components in the order in which they are first used in the course of replication: helicase, single-strand-binding protein, DNA gyrase, initiator protein.
Elongation
In the elongation phase of replication, single-stranded DNA is used as a template for the synthesis of DNA. This process requires a series of enzymes.
The Synthesis of Primers
All DNA polymerases require a nucleotide with a 3′-OH group to which a new nucleotide can be added. Because of this requirement, DNA polymerases cannot initiate DNA synthesis on a bare template; rather, they require a primer—an existing 3′-OH group—to get started. How, then, does DNA synthesis begin?
An enzyme called primase synthesizes short stretches (about 10-12 nucleotides long) of RNA nucleotides, or primers, which provide a 3′-OH group to which DNA polymerase can attach DNA nucleotides. (Because primase is an RNA polymerase, it does not require a pre-existing 3′-OH group to start synthesis of a nucleotide strand.) All DNA molecules initially have short RNA primers embedded within them; these primers are later removed and replaced with DNA nucleotides.
On the leading strand, where DNA synthesis is continuous, a primer is required only at the 5′ end of the newly synthesized strand. On the lagging strand, where replication is discontinuous, a new primer must be generated at the beginning of each Okazaki fragment (Figure 12.13). Primase forms a complex with helicase at the replication fork and moves along the template of the lagging strand. The single primer on the leading strand is probably synthesized by the primase–helicase complex on the template of the lagging strand of the other replication fork, at the opposite end of the replication bubble.  TRY PROBLEM 30
TRY PROBLEM 30
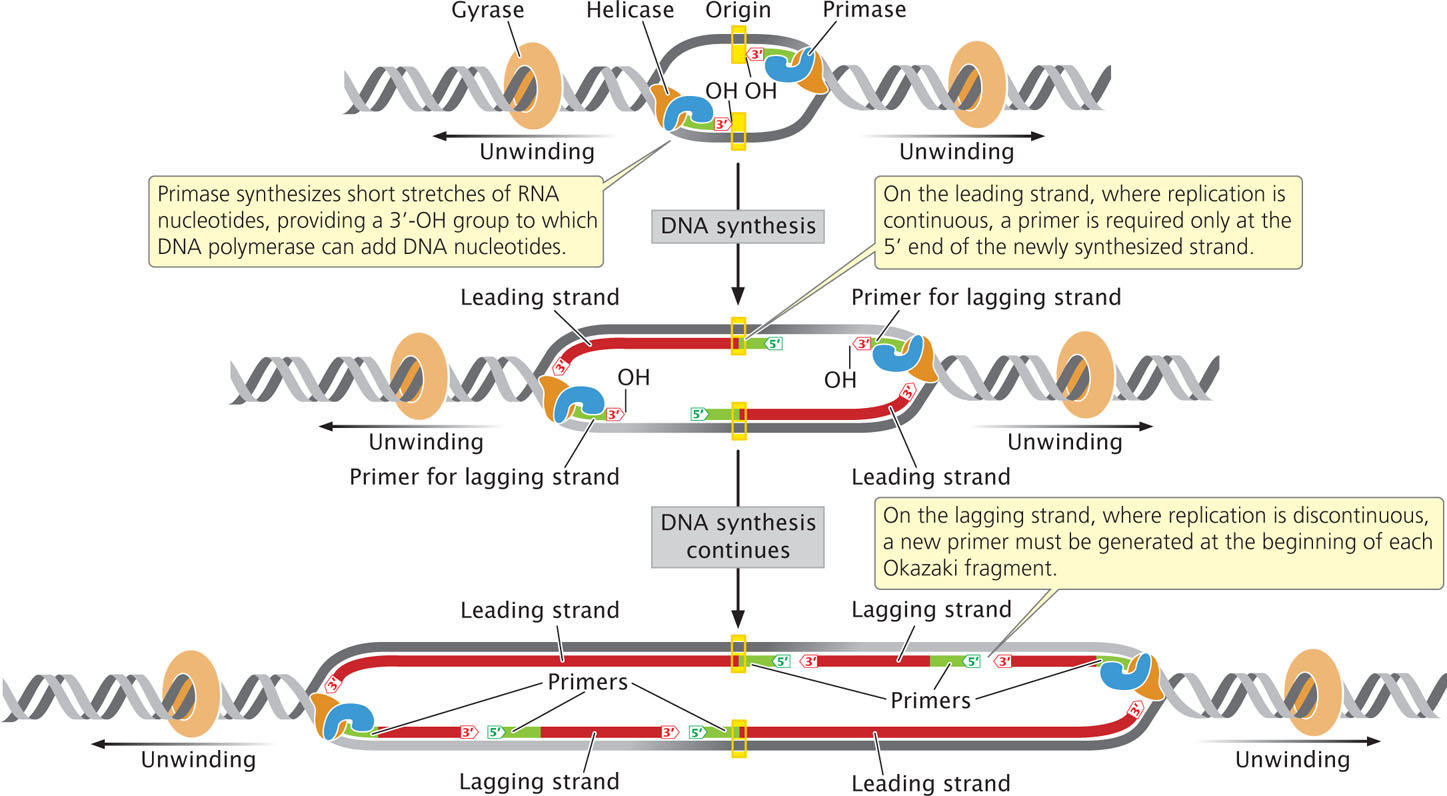
CONCEPTS
Primase synthesizes a short stretch of RNA nucleotides (primers), which provides a 3′-OH group for the attachment of DNA nucleotides to start DNA synthesis.
 CONCEPT CHECK 5
CONCEPT CHECK 5Primers are synthesized where on the lagging strand?
- Only at the 5′ end of the newly synthesized strand
- Only at the 3′ end of the newly synthesized strand
- At the beginning of every Okazaki fragment
- At multiple places within an Okazaki fragment
DNA Synthesis By DNA Polymerases
After DNA has unwound and a primer has been added, DNA polymerases elongate the new polynucleotide strand by catalyzing DNA polymerization. The best-studied polymerases are those of E. coli, which has at least five different DNA polymerases. Two of them, DNA polymerase I and DNA polymerase III, carry out DNA synthesis in replication (Table 12.3); the other three have specialized functions in DNA repair.
| DNA Polymerase | 5′→3′ Polymerization | 3′→5′ Exonuclease | 5′→3′ Exonuclease | Function |
|---|---|---|---|---|
| I | Yes | Yes | Yes | Removes and replaces primers |
| II | Yes | Yes | No | DNA repair; restarts replication after damaged DNA halts synthesis |
| III | Yes | Yes | No | Elongates DNA |
| IV | Yes | No | No | DNA repair |
| V | Yes | No | No | DNA repair; translesion DNA synthesis |
DNA polymerase III is a large multiprotein complex that acts as the main workhorse of replication. DNA polymerase III synthesizes nucleotide strands by adding new nucleotides to the 3′ end of a growing DNA molecule. This enzyme has two enzymatic activities (see Table 12.3). Its 5′→3′ polymerase activity allows it to add new nucleotides in the 5′→3′ direction. Its 3′→5′ exonuclease activity allows it to remove nucleotides in the 3′→5′ direction, enabling it to correct errors. If a nucleotide having an incorrect base is inserted into the growing DNA molecule, DNA polymerase III uses its 3′→5′ exonuclease activity to back up and remove the incorrect nucleotide. It then resumes its 5′→3′ polymerase activity. These two functions together allow DNA polymerase III to efficiently and accurately synthesize new DNA molecules. DNA polymerase III has high processivity, which means that it is capable of adding many nucleotides to the growing DNA strand without releasing the template: it normally holds on to the template and continues synthesizing DNA until the template has been completely replicated. The high processivity of DNA polymerase III is ensured by one of the polypeptides that constitutes the enzyme. This polypeptide, termed the β subunit, serves as a clamp for the polymerase enzyme: it encircles the DNA and keeps the DNA polymerase attached to the template strand during replication. DNA polymerase III adds DNA nucleotides to the primer, synthesizing the DNA of both the leading and the lagging strands.
The first E. coli polymerase to be discovered, DNA polymerase I, also has 5′→3′ polymerase and 3′→5′ exonuclease activities (see Table 12.3), permitting the enzyme to synthesize DNA and to correct errors. Unlike DNA polymerase III, however, DNA polymerase I also possesses 5′→3′ exonuclease activity, which is used to remove the primers laid down by primase and to replace them with DNA nucleotides by synthesizing in a 5′→3′ direction. DNA polymerase I has lower processivity than DNA polymerase III. The removal and replacement of primers appear to constitute the main function of DNA polymerase I. After DNA polymerase III has initiated synthesis at the primer and moved downstream, DNA polymerase I removes the RNA nucleotides of the primer, replacing them with DNA nucleotides. DNA polymerases II, IV, and V function in DNA repair.
Despite their differences, all of E. coli’s DNA polymerases
- 1. synthesize any sequence specified by the template strand;
- 2. synthesize in the 5′→3′ direction by adding nucleotides to a 3′-OH group;
- 3. use dNTPs to synthesize new DNA;
- 4. require a 3′ OH-group to initiate synthesis;
- 5. catalyze the formation of a phosphodiester bond by joining the 5’-phosphate group of the incoming nucleotide to the 3′-OH group of the preceding nucleotide on the growing strand, cleaving off two phosphates in the process;
- 6. produce newly synthesized strands that are complementary and antiparallel to the template strands;
- 7. and are associated with a number of other proteins.
 TRY PROBLEM 27
TRY PROBLEM 27
CONCEPTS
DNA polymerases synthesize DNA in the 5′→3′ direction by adding new nucleotides to the 3′ end of a growing nucleotide strand.
DNA Ligase
After DNA polymerase III attaches a DNA nucleotide to the 3′-OH group on the last nucleotide of the RNA primer, each new DNA nucleotide then provides the 3′-OH group needed for the next DNA nucleotide to be added. This process continues as long as a template is available (Figure 12.14a). DNA polymerase I follows DNA polymerase III and, using its 5′→3′ exonuclease activity, removes the RNA primer. It then uses its 5′→3′ polymerase activity to replace the RNA nucleotides with DNA nucleotides. DNA polymerase I attaches the first nucleotide to the OH group at the 3′ end of the preceding Okazaki fragment and then continues, in the 5′→3′ direction along the nucleotide strand, removing and replacing, one at a time, the RNA nucleotides of the primer (Figure 12.14b).
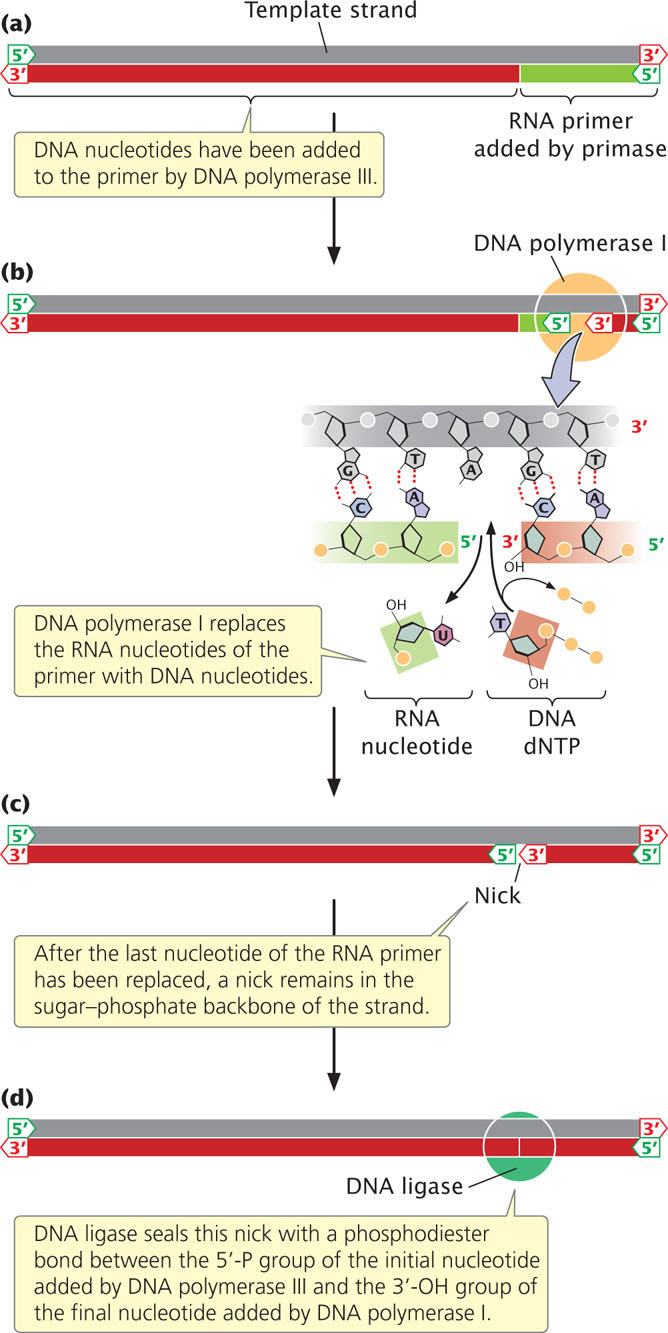
After polymerase I has replaced the last nucleotide of the RNA primer with a DNA nucleotide, a break remains in the sugar–phosphate backbone of the new DNA strand. The 3′-OH group of the last nucleotide to have been added by DNA polymerase I is not attached to the 5′-phosphate group of the first nucleotide added by DNA polymerase III (Figure 12.14c). This break is sealed by the enzyme DNA ligase, which catalyzes the formation of a phosphodiester bond without adding another nucleotide to the strand (Figure 12.14d). Some of the major enzymes and proteins required for prokaryotic DNA replication are summarized in Table 12.4.
| Component | Function |
|---|---|
| Initiator protein | Binds to origin and separates strands of DNA to initiate replication |
| DNA helicase | Unwinds DNA at replication fork |
| Single-strand-binding proteins | Attach to single-stranded DNA and prevent secondary structures from forming |
| DNA gyrase | Moves ahead of the replication fork, making and resealing breaks in the double-helical DNA to release the torque that builds up as a result of unwinding at the replication fork |
| DNA primase | Synthesizes a short RNA primer to provide a 3′-OH group for the attachment of DNA nucleotides |
| DNA polymerase III | Elongates a new nucleotide strand from the 3′-OH group provided by the primer |
| DNA polymerase I | Removes RNA primers and replaces them with DNA |
| DNA ligase | Joins Okazaki fragments by sealing breaks in the sugar–phosphate backbone of newly synthesized DNA |
CONCEPTS
After primers have been removed and replaced, the break in the sugar–phosphate linkage is sealed by DNA ligase.
 CONCEPT CHECK 6
CONCEPT CHECK 6Which bacterial enzyme removes the primers?
- Primase
- DNA polymerase I
- DNA polymerase III
- Ligase
Elongation at The Replication Fork
Now that the major enzymatic components of elongation—DNA polymerases, helicase, primase, and ligase—have been introduced, let’s consider how these components interact at the replication fork. Because the synthesis of both strands takes place simultaneously, two units of DNA polymerase III must be present at the replication fork, one for each strand. In one model of the replication process, the two units of DNA polymerase III are connected (Figure 12.15); the lagging-strand template loops around so that it is in position for 5′→3′ replication. In this way, the DNA polymerase III complex is able to carry out 5′→3′ replication simultaneously on both templates, even though they run in opposite directions. After about 1000 bp of new DNA has been synthesized, DNA polymerase III releases the lagging-strand template, and a new loop forms (see Figure 12.15). Primase synthesizes a new primer on the lagging strand and DNA polymerase III then synthesizes a new Okazaki fragment. See how replication takes place on both strands simultaneously by viewing  Animation 12.2.
Animation 12.2.
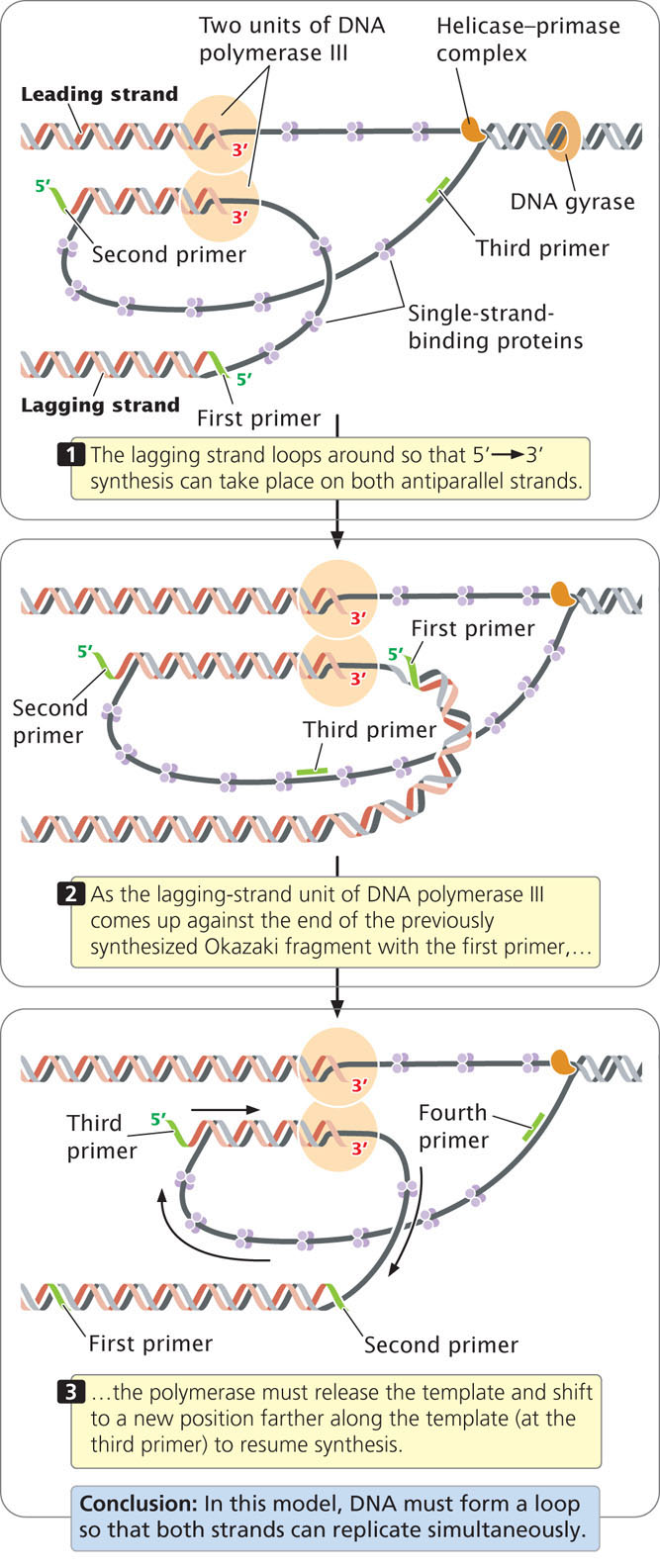
In summary, each active replication fork requires five basic components:
- 1. helicase to unwind the DNA,
- 2. single-strand-binding proteins to protect the single nucleotide strands and prevent secondary structures,
- 3. the topoisomerase gyrase to remove strain ahead of the replication fork,
- 4. primase to synthesize primers with a 3′-OH group at the beginning of each DNA fragment, and
- 5. DNA polymerase to synthesize the leading and lagging nucleotide strands.
 You can see how the different components of the replication process work together by viewing Animations 12.3 and 12.4.
You can see how the different components of the replication process work together by viewing Animations 12.3 and 12.4.
Termination
In some DNA molecules, replication is terminated whenever two replication forks meet. In others, specific termination sequences (called Ter sites) block further replication. A termination protein, called Tus in E. coli, binds to these sequences, creating a Tus-Ter complex that blocks the movement of helicase, thus stalling the replication fork and preventing further DNA replication. Each Tus-Ter complex blocks a replication fork moving in one direction but not the other.
The Fidelity of DNA Replication
Overall, the error rate in replication is less than one mistake per billion nucleotides. How is this incredible accuracy achieved?
DNA polymerases are very particular in pairing nucleotides with their complements on the template strand. Errors in nucleotide selection by DNA polymerase arise only about once per 100,000 nucleotides. Most of the errors that do arise in nucleotide selection are corrected in a second process called proofreading. When a DNA polymerase inserts an incorrect nucleotide into the growing strand, the 3′-OH group of the mispaired nucleotide is not correctly positioned in the active site of the DNA polymerase for accepting the next nucleotide. The incorrect positioning stalls the polymerization reaction, and the 3′→5′ exonuclease activity of DNA polymerase removes the incorrectly paired nucleotide. DNA polymerase then inserts the correct nucleotide. Together, proofreading and nucleotide selection result in an error rate of only one in 10 million nucleotides.
A third process, called mismatch repair (discussed further in Chapter 18), corrects errors after replication is complete. Any incorrectly paired nucleotides remaining after replication produce a deformity in the secondary structure of the DNA; the deformity is recognized by enzymes that excise an incorrectly paired nucleotide and use the original nucleotide strand as a template to replace the incorrect nucleotide. Mismatch repair requires the ability to distinguish between the old and the new strands of DNA, because the enzymes need some way of determining which of the two incorrectly paired bases to remove. In E. coli, methyl groups (—CH3) are added to particular nucleotide sequences, but only after replication. Thus, immediately after DNA synthesis, only the old DNA strand is methylated. It can therefore be distinguished from the newly synthesized strand, and mismatch repair takes place preferentially on the unmethylated nucleotide strand. No single process could produce this level of accuracy; a series of processes are required, each process catching errors missed by the preceding ones.
CONCEPTS
Replication is extremely accurate, with less than one error per billion nucleotides. The high level of accuracy in DNA replication is produced by nucleotide selection, proofreading, and mismatch repair.
 CONCEPT CHECK 7
CONCEPT CHECK 7Which mechanism requires the ability to distinguish between newly synthesized and template strands of DNA?
- Nucleotide selection
- DNA proofreading
- Mismatch repair
- All of the above
CONNECTING CONCEPTS
Bacterial replication requires a number of enzymes (see Table 12.4), proteins, and DNA sequences that function together to synthesize a new DNA molecule. These components are important, but we must not become so immersed in the details of the process that we lose sight of the general principles of replication.
- 1. Replication is always semiconservative.
- 2. Replication begins at sequences called origins.
- 3. DNA synthesis is initiated by short segments of RNA called primers.
- 4. The elongation of DNA strands is always in the 5′➝ 3′ direction.
- 5. New DNA is synthesized from dNTPs; in the polymerization of DNA, two phosphate groups are cleaved from a dNTP and the resulting nucleotide is added to the 3′-OH group of the growing nucleotide strand.
- 6. Replication is continuous on the leading strand and discontinuous on the lagging strand.
- 7. New nucleotide strands are complementary and antiparallel to their template strands.
- 8. Replication takes place at very high rates and is astonishingly accurate, thanks to precise nucleotide selection, proofreading, and mismatch repair.