16.2 Operons Control Transcription in Bacterial Cells
A significant difference between bacterial and eukaryotic gene control lies in the organization of functionally related genes. As discussed in the introduction to this chapter, many bacterial genes that have related functions are clustered and under the control of a single promoter. These genes are often transcribed together into a single mRNA. A group of bacterial structural genes that are transcribed together (along with their promoter and additional sequences that control transcription) is called an operon. The operon regulates the expression of the structural genes by controlling transcription, which, in bacteria, is usually the most important level of gene regulation.
Operon Structure
The organization of a typical operon is illustrated in Figure 16.3. At one end of the operon is a set of structural genes, shown in Figure 16.3 as gene a, gene b, and gene c. These structural genes are transcribed into a single mRNA, which is translated to produce enzymes A, B, and C. These enzymes carry out a series of biochemical reactions that convert precursor molecule X into product Y. The transcription of structural genes a, b, and c is under the control of a promoter, which lies upstream of the first structural gene. RNA polymerase binds to the promoter and then moves downstream, transcribing the structural genes.
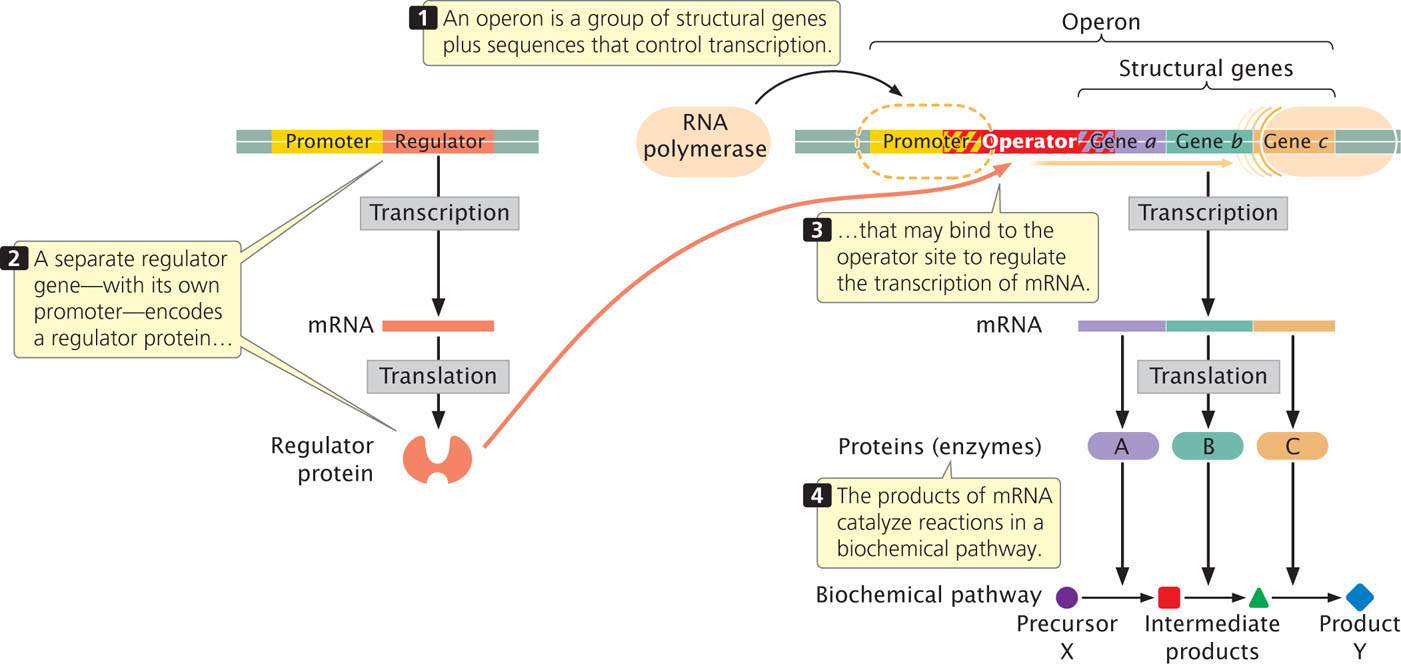
A regulator gene helps to control the transcription of the structural genes of the operon. Although it affects operon function, the regulator gene is not considered part of the operon. The regulator gene has its own promoter and is transcribed into a short mRNA, which is translated into a small protein. This regulator protein can bind to a region of the operon called the operator and affect whether transcription can take place. The operator usually overlaps the 3′ end of the promoter and sometimes the 5′ end of the first structural gene (see Figure 16.3).
CONCEPTS
Functionally related genes in bacterial cells are frequently clustered together as a single transcriptional unit termed an operon. A typical operon includes several structural genes, a promoter for the structural genes, and an operator site to which the product of a regulator gene binds.
 CONCEPT CHECK 4
CONCEPT CHECK 4What is the difference between a structural gene and a regulator gene?
- Structural genes are transcribed into mRNA, but regulator genes aren’t.
- Structural genes have complex structures; regulator genes have simple structures.
- Structural genes encode proteins that function in the structure of the cell; regulator genes carry out metabolic reactions.
- Structural genes encode proteins; regulator genes control the transcription of structural genes.
Negative and Positive Control: Inducible and Repressible Operons
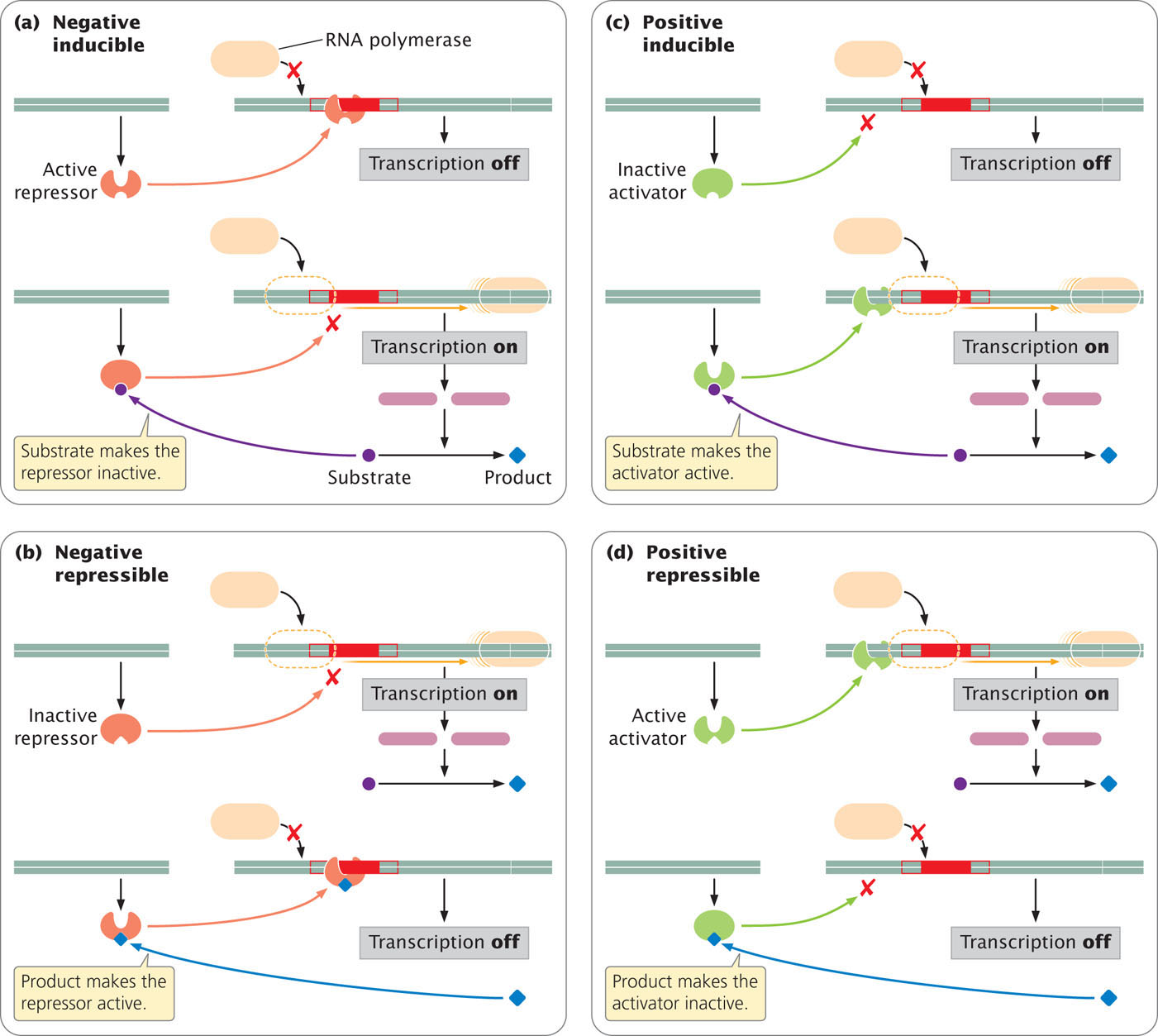
There are two types of transcriptional control: negative control, in which a regulatory protein is a repressor, binding to DNA and inhibiting transcription; and positive control, in which a regulatory protein is an activator, stimulating transcription. Operons can also be either inducible or repressible. Inducible operons are those in which transcription is normally off (not taking place); something must happen to induce transcription, or turn it on. Repressible operons are those in which transcription is normally on (taking place); something must happen to repress transcription, or turn it off. In the next sections, we will consider several varieties of these basic control mechanisms.
Negative Inducible Operons
In a negative inducible operon, the regulator gene encodes an active repressor that readily binds to the operator (Figure 16.4a). Because the operator site overlaps the promoter site, the binding of this protein to the operator physically blocks the binding of RNA polymerase to the promoter and prevents transcription. For transcription to take place, something must happen to prevent the binding of the repressor at the operator site. This type of system is said to be inducible because transcription is normally off (inhibited) and must be turned on (induced).
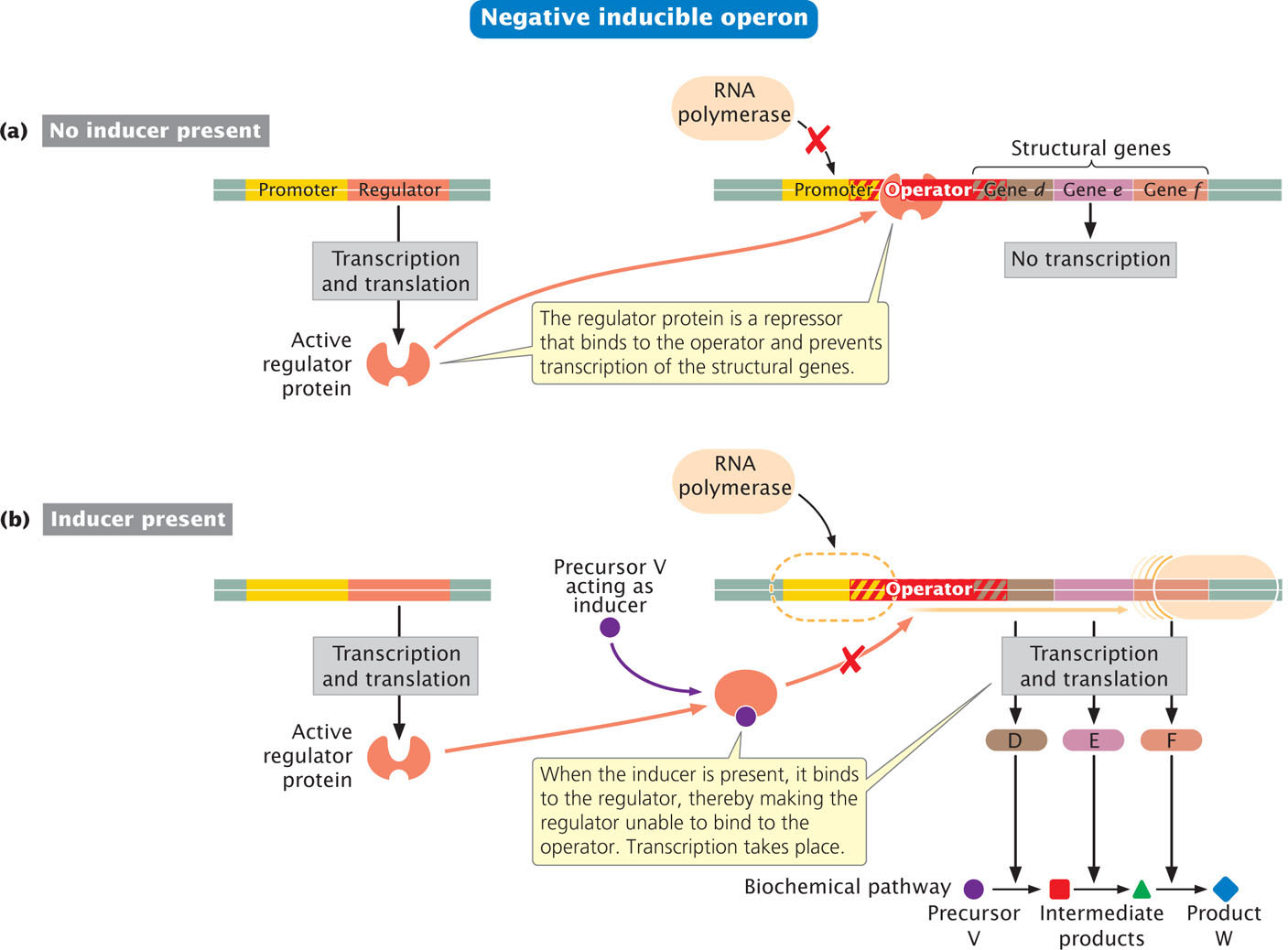
Transcription is turned on when a small molecule, an inducer, binds to the repressor (Figure 16.4b). Regulatory proteins frequently have two binding sites: one that binds to DNA and another that binds to a small molecule such as an inducer. The binding of the inducer (precursor V in Figure 16.4b) alters the shape of the repressor, preventing it from binding to DNA. Proteins of this type, which change shape on binding to another molecule, are called allosteric proteins.
When the inducer is absent, the repressor binds to the operator, the structural genes are not transcribed, and enzymes D, E, and F (which metabolize precursor V) are not synthesized (see Figure 16.4a). This mechanism is an adaptive one: because no precursor V is available, synthesis of the enzymes would be wasteful when they have no substrate to metabolize. As soon as precursor V becomes available, some of it binds to the repressor, rendering the repressor inactive and unable to bind to the operator site. RNA polymerase can now bind to the promoter and transcribe the structural genes. The resulting mRNA is then translated into enzymes D, E, and F, which convert substrate V into product W (see Figure 16.4b). So, an operon with negative inducible control regulates the synthesis of the enzymes economically: the enzymes are synthesized only when their substrate (V) is available.
Inducible operons usually control proteins that carry out degradative processes—proteins that break down molecules. For these types of proteins, inducible control makes sense because the proteins are not needed unless the substrate (which is broken down by the proteins) is present.
Negative Repressible Operons
Some operons with negative control are repressible, meaning that transcription normally takes place and must be turned off, or repressed. The regulator protein in this type of operon also is a repressor but is synthesized in an inactive form that cannot by itself bind to the operator. Because no repressor is bound to the operator, RNA polymerase readily binds to the promoter and transcription of the structural genes takes place (Figure 16.5a).
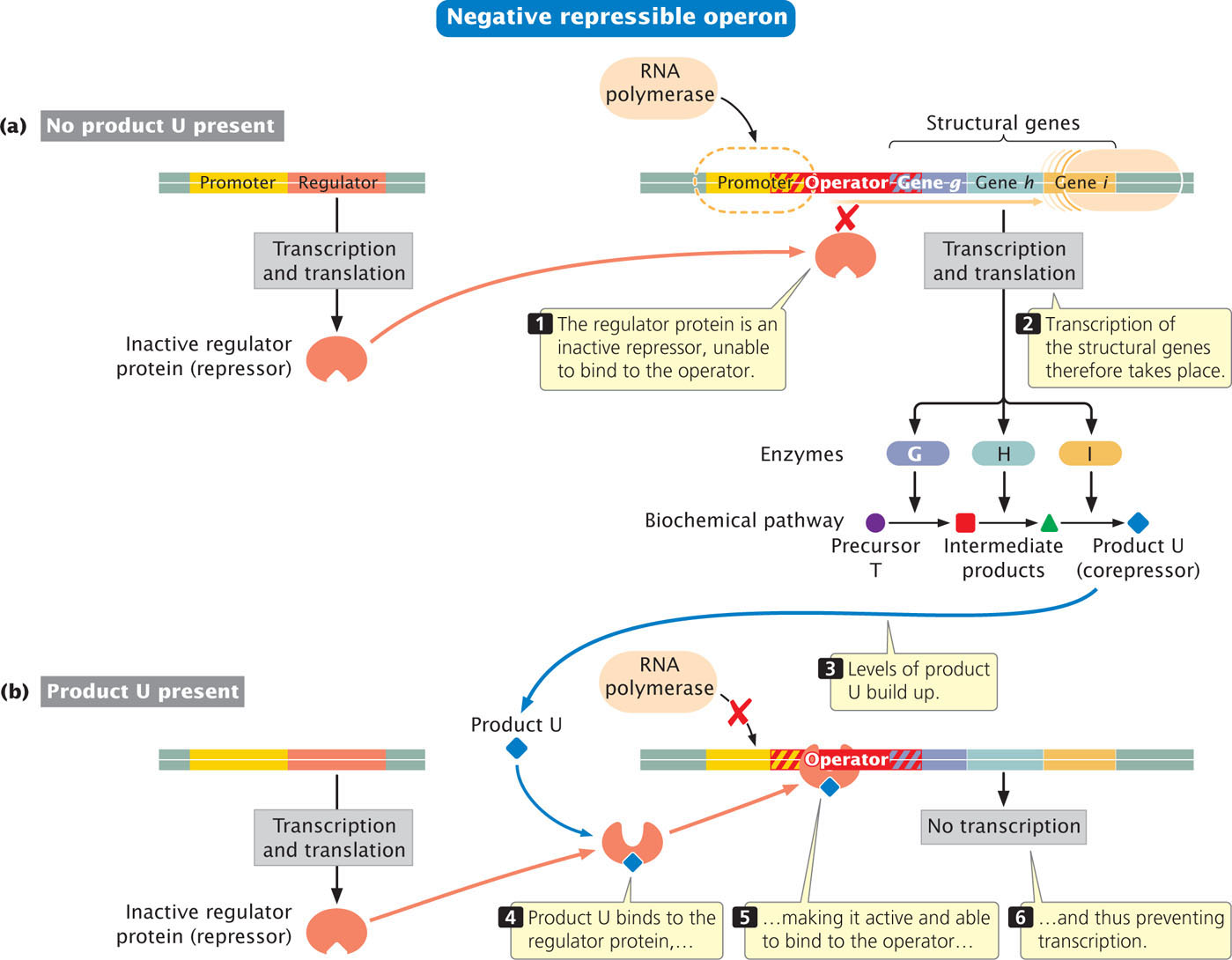
To turn transcription off, something must happen to make the repressor active. A small molecule called a corepressor binds to the repressor and makes it capable of binding to the operator. In the example illustrated (see Figure 16.5a), the product (U) of the metabolic reaction is the corepressor. As long as the level of product U is high, it is available to bind to the repressor and activate it, preventing transcription (Figure 16.5b). With the operon repressed, enzymes G, H, and I are not synthesized, and no more U is produced from precursor T. However, when all of product U is used up, the repressor is no longer activated by product U and cannot bind to the operator. The inactivation of the repressor allows the transcription of the structural genes and the synthesis of enzymes G, H, and I, resulting in the conversion of precursor T into product U. Like inducible operons, repressible operons are economical: the enzymes are synthesized only as needed.
Repressible operons usually control proteins that carry out the biosynthesis of molecules needed in the cell, such as amino acids. For these types of operons, repressible control makes sense because the product produced by the proteins is always needed by the cell. Thus, these operons are normally on and are turned off when there are adequate amounts of the product already present.
Note that both the inducible and the repressible systems that we have considered are forms of negative control, in which the regulatory protein is a repressor. We will now consider positive control, in which a regulator protein stimulates transcription.
Positive Control
With positive control, a regulatory protein is an activator: it binds to DNA (usually at a site other than the operator) and stimulates transcription. Positive control can be inducible or repressible.
In a positive inducible operon, transcription is normally turned off because the regulator protein (an activator) is produced in an inactive form. Transcription takes place when an inducer has become attached to the regulatory protein, rendering the regulator active. Logically, the inducer should be the precursor of the reaction controlled by the operon so that the necessary enzymes would be synthesized only when the substrate for their reaction was present.
A positive operon can also be repressible; the regulatory protein is produced in a form that readily binds to DNA, meaning that transcription normally takes place and has to be repressed. Transcription is inhibited when a substance becomes attached to the activator and renders it unable to bind to the DNA so that transcription is no longer stimulated. Here, the product (P) of the reaction controlled by the operon would logically be the repressing substance, because it would be economical for the cell to prevent the transcription of genes that allow the synthesis of P when plenty of P was already available. The characteristics of positive and negative control in inducible and repressible operons are summarized in Figure 16.6.  TRY PROBLEM 11
TRY PROBLEM 11
CONCEPTS
There are two basic types of transcriptional control: negative and positive. In negative control, when a regulatory protein (repressor) binds to DNA, transcription is inhibited; in positive control, when a regulatory protein (activator) binds to DNA, transcription is stimulated. Some operons are inducible; transcription is normally off and must be turned on. Other operons are repressible; transcription is normally on and must be turned off.
 CONCEPT CHECK 5
CONCEPT CHECK 5In a negative repressible operon, the regulator protein is synthesized as
- an active activator.
- an inactive activator.
- an active repressor.
- an inactive repressor.
The lac Operon of E. coli
In 1961, François Jacob and Jacques Monod described the “operon model” for the genetic control of lactose metabolism in E. coli. This work and subsequent research on the genetics of lactose metabolism established the operon as the basic unit of transcriptional control in bacteria. Despite the fact that, at the time, no methods were available for determining nucleotide sequences, Jacob and Monod deduced the structure of the operon genetically by analyzing the interactions of mutations that interfered with the normal regulation of lactose metabolism. We will examine the effects of some of these mutations after seeing how the lac operon regulates lactose metabolism.
Lactose Metabolism
Lactose is a major carbohydrate found in milk; it can be metabolized by E. coli bacteria that reside in the mammalian gut. Lactose does not easily diffuse across the E. coli cell membrane and must be actively transported into the cell by the protein permease (Figure 16.7). To utilize lactose as an energy source, E. coli must first break it into glucose and galactose, a reaction catalyzed by the enzyme β-galactosidase. This enzyme can also convert lactose into allolactose, a compound that plays an important role in regulating lactose metabolism. A third enzyme, thiogalactoside transacetylase, is also produced by the lac operon, but its function in lactose metabolism is not yet clear. One possible function is detoxification, preventing the accumulation of thiogalactosides that are transported into the cell along with lactose by lactose permease.
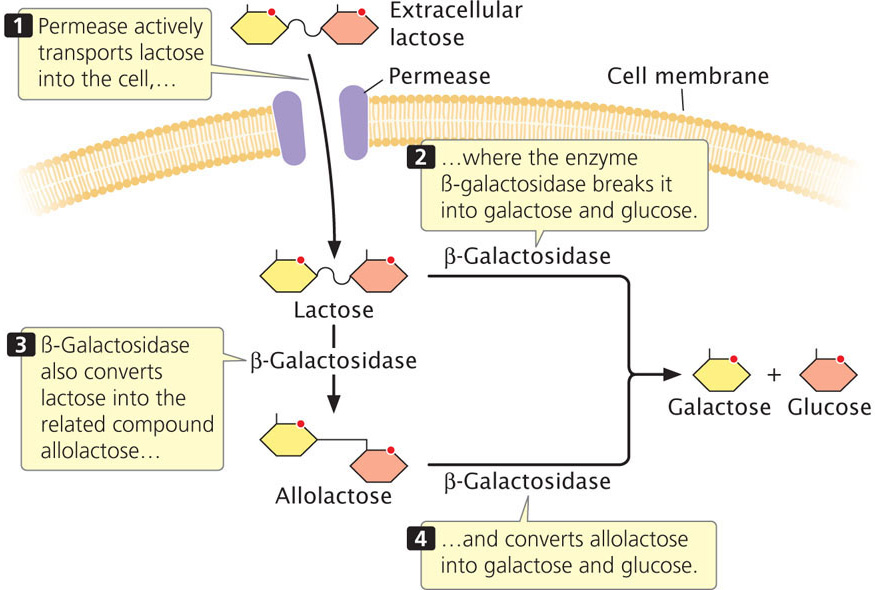
Regulation of the Lac Operon
The lac operon is an example of a negative inducible operon. The enzymes β-galactosidase, permease, and transacetylase are encoded by adjacent structural genes in the lac operon of E. coli (Figure 16.8a) and have a common promoter (lacP in Figure 16.8a). β-Galactosidase is encoded by the lacZ gene, permease by the lacY gene, and transacetylase by the lacA gene. When lactose is absent from the medium in which E. coli grows, few molecules of each protein are produced. If lactose is added to the medium and glucose is absent, the rate of synthesis of all three proteins simultaneously increases about a thousandfold within 2 to 3 minutes. This boost in protein synthesis results from the transcription of lacZ, lacY, and lacA and exemplifies coordinate induction, the simultaneous synthesis of several proteins, stimulated by a specific molecule, the inducer (Figure 16.8b).
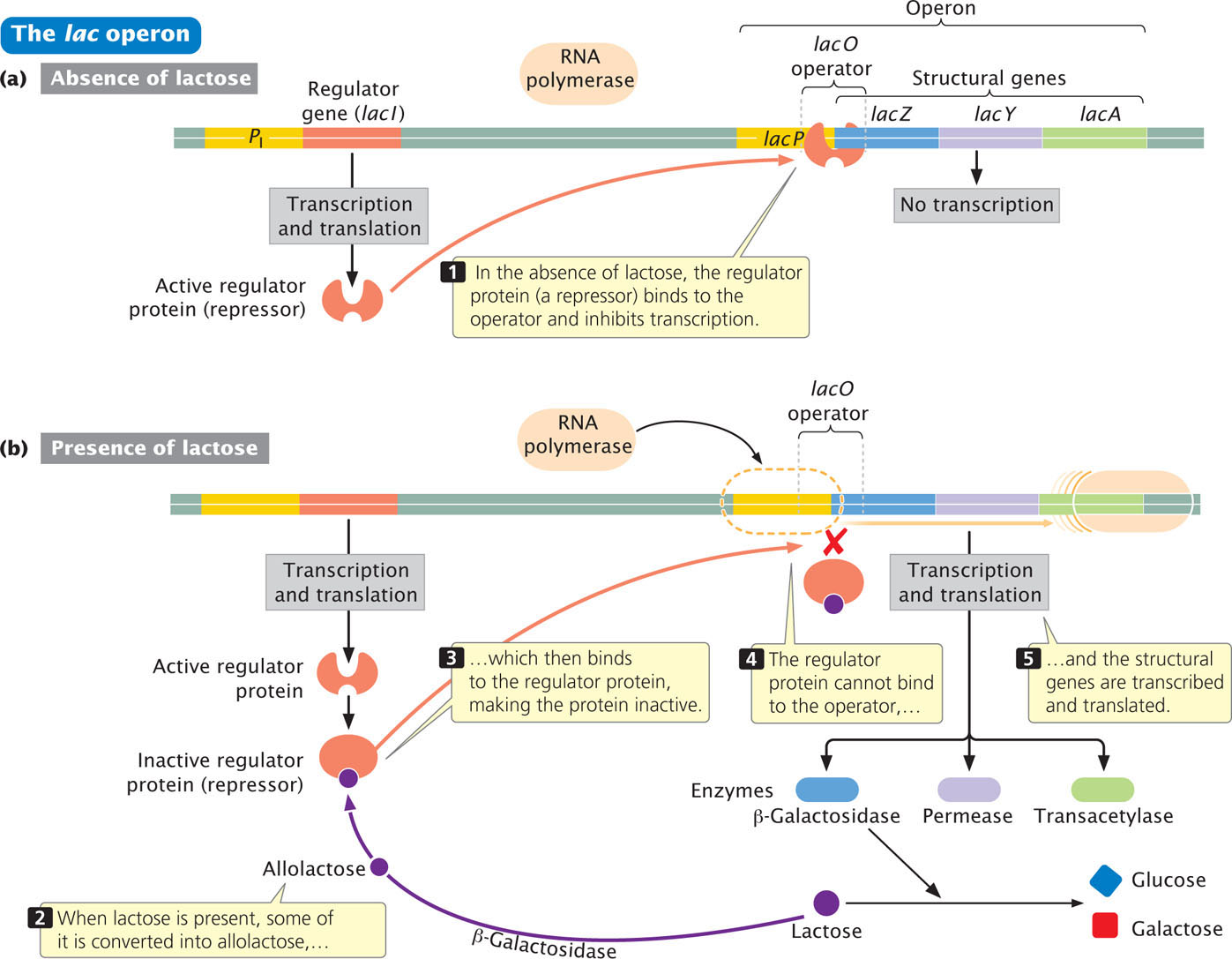
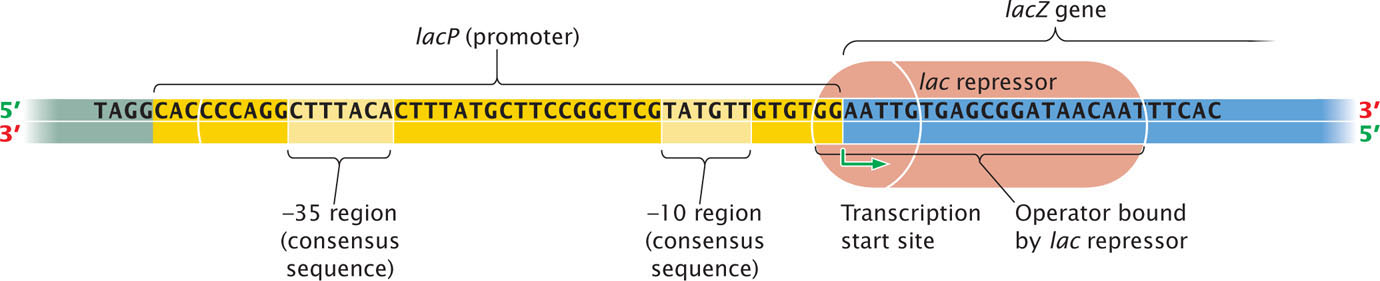
Although lactose appears to be the inducer here, allolactose is actually responsible for induction. Upstream of lacP is a regulator gene, lacI, which has its own promoter (PI). The lacI gene is transcribed into a short mRNA that is translated into a repressor. The repressor consists of four identical polypeptides and has two types of binding sites; one type of site binds to allolactose and the other binds to DNA. In the absence of lactose (and, therefore, allolactose), the repressor binds to the lac operator site lacO (see Figure 16.8a). Jacob and Monod mapped the operator to a position adjacent to the lacZ gene; more-recent nucleotide sequencing has demonstrated that the operator actually overlaps the 3′ end of the promoter and the 5′ end of lacZ (Figure 16.9)
RNA polymerase binds to the promoter and moves down the DNA molecule, transcribing the structural genes. When the repressor is bound to the operator, the binding of RNA polymerase is blocked, and transcription is prevented. When lactose is present, some of it is converted into allolactose, which binds to the repressor and causes the repressor to be released from the DNA. In the presence of lactose, then, the repressor is inactivated, the binding of RNA polymerase is no longer blocked, the transcription of lacZ, lacY, and lacA takes place, and the lac proteins are produced.
Have you spotted the flaw in the explanation just given for the induction of the lac proteins? You might recall that permease is required to transport lactose into the cell. If the lac operon is repressed and no permease is being produced, how does lactose get into the cell to inactivate the repressor and turn on transcription? Furthermore, the inducer is actually allolactose, which must be produced from lactose by β-galactosidase. If β-galactosidase production is repressed, how can lactose metabolism be induced?
The answer is that repression never completely shuts down transcription of the lac operon. Even with active repressor bound to the operator, there is a low level of transcription and a few molecules of β-galactosidase, permease, and transacetylase are synthesized. When lactose appears in the medium, the permease that is present transports a small amount of lactose into the cell. There, the few molecules of β-galactosidase that are present convert some of the lactose into allolactose, which then induces transcription.
Several compounds related to allolactose also can bind to the lac repressor and induce transcription of the lac operon. One such inducer is isopropylthiogalactoside (IPTG). Although IPTG inactivates the repressor and allows the transcription of lacZ, lacY, and lacA, this inducer is not metabolized by β-galactosidase; for this reason, IPTG is often used in research to examine the effects of induction, independent of metabolism.
CONCEPTS
The lac operon of E. coli controls the transcription of three genes needed in lactose metabolism: the lacZ gene, which encodes β-galactosidase; the lacY gene, which encodes permease; and the lacA gene, which encodes thiogalactoside transacetylase. The lac operon is negative inducible: a regulator gene produces a repressor that binds to the operator site and prevents the transcription of the structural genes. The presence of allolactose inactivates the repressor and allows the transcription of the lac operon.
 CONCEPT CHECK 6
CONCEPT CHECK 6In the presence of allolactose, the lac repressor
- binds to the operator.
- binds to the promoter.
- cannot bind to the operator.
- binds to the regulator gene.
lac Mutations
Jacob and Monod worked out the structure and function of the lac operon by analyzing mutations that affected lactose metabolism. To help define the roles of the different components of the operon, they used partial diploid strains of E. coli. The cells of these strains possessed two different DNA molecules: the full bacterial chromosome and an extra piece of DNA. Jacob and Monod created these strains by allowing conjugation to take place between two bacteria (see Chapter 9). In conjugation, a small circular piece of DNA (the F plasmid, see Chapter 9) is transferred from one bacterium to another. The F plasmid used by Jacob and Monod contained the lac operon so the recipient bacterium became partly diploid, possessing two copies of the lac operon. By using different combinations of mutations on the bacterial and plasmid DNA, Jacob and Monod determined that some parts of the lac operon are cis acting (able to control the expression of genes only when on the same piece of DNA), whereas other parts are trans acting (able to control the expression of genes on other DNA molecules).
Structural-Gene Mutations
Jacob and Monod first discovered some mutant strains that had lost the ability to synthesize either β-galactosidase or permease (they did not study in detail the effects of mutations on the transacetylase enzyme, and so transacetylase will not be considered here). The mutations in the mutant strains mapped to the lacZ or lacY structural genes and altered the amino acid sequences of the proteins encoded by the genes. These mutations clearly affected the structure of the proteins but not the regulation of their synthesis.
Through the use of partial diploids, Jacob and Monod were able to establish that mutations at the lacZ and lacY genes were independent and usually affected only the product of the gene in which the mutation occurred. Partial diploids with lacZ+ lacY− on the bacterial chromosome and lacZ− lacY+ on the plasmid functioned normally, producing β-galactosidase and permease in the presence of lactose. (The genotype of a partial diploid is written by separating the genes on each DNA molecule with a slash: lacZ+ lacY−/lacZ− lacY+.) In this partial diploid, a single functional β-galactosidase gene (lacZ+) is sufficient to produce β-galactosidase; whether the functional β-galactosidase gene is coupled to a functional (lacY+) or a defective (lacY−) permease gene makes no difference. The same is true of the lacY+ gene.
Regulator-Gene Mutations
Jacob and Monod also isolated mutations that affected the regulation of protein production. Mutations in the lacI gene affect the production of both β-galactosidase and permease because genes for both proteins are in the same operon and are regulated coordinately.
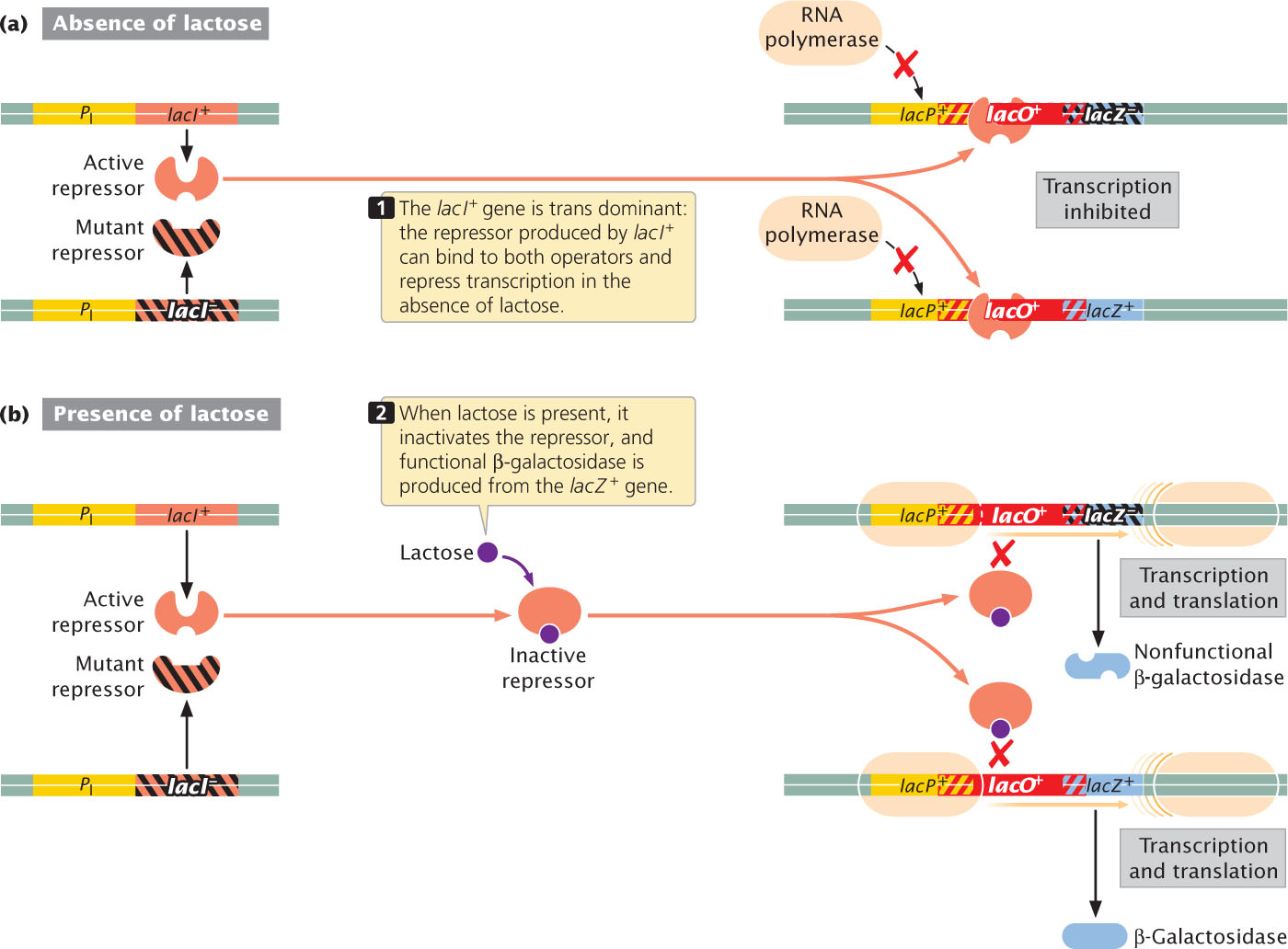
Some of these mutations were constitutive, causing the lac proteins to be produced all the time, whether lactose was present or not. Such mutations in the regulator gene were designated lacI−. The construction of partial diploids demonstrated that a lacI+ gene is dominant over a lacI− gene; a single copy of lacI+ (genotype lacI+/lacI− was sufficient to bring about normal regulation of protein production. Furthermore, lacI+ restored normal control to an operon even if the operon was located on a different DNA molecule, showing that lacI+ can be trans acting. A partial diploid with genotype lacI+ lacZ−/lacI− lacZ+ functioned normally, synthesizing β-galactosidase only when lactose was present (Figure 16.10). In this strain, the lacI+ gene on the bacterial chromosome was functional, but the lacZ− gene was defective; on the plasmid, the lacI− gene was defective, but the lacZ+ gene was functional. The fact that a lacI+ gene could regulate a lacZ+ gene located on a different DNA molecule indicated to Jacob and Monod that the lacI+ gene product was able to operate on either the plasmid or the chromosome.
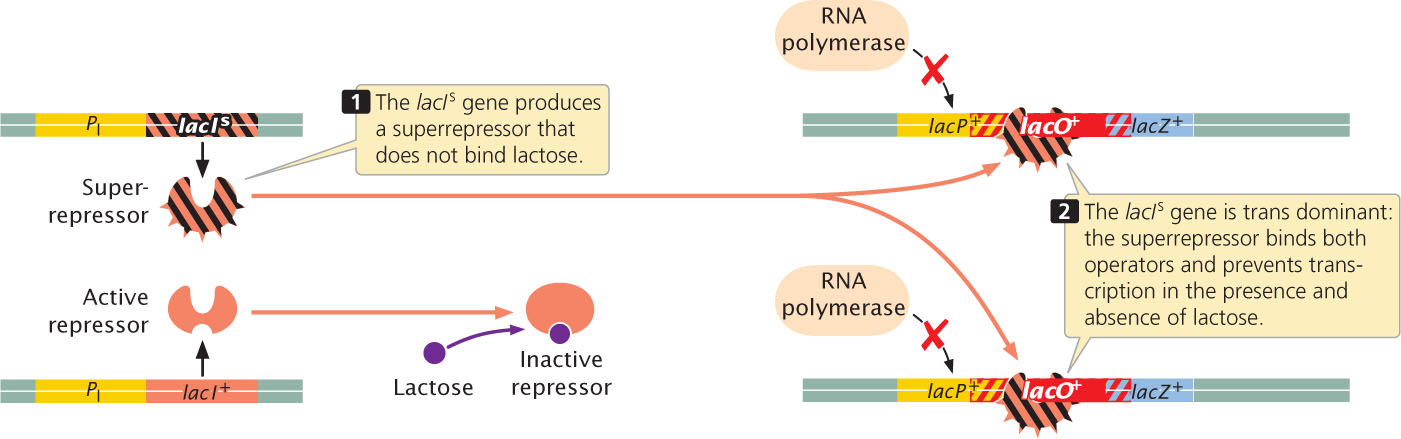
Some lacI mutations isolated by Jacob and Monod prevented transcription from taking place even in the presence of lactose. These mutations were referred to as superrepressors (lacIs), because they produced defective repressors that could not be inactivated by an inducer. The lacIs mutations produced a repressor with an altered inducer-binding site, which made the inducer unable to bind to the repressor; consequently, the repressor was always able to attach to the operator site and prevent transcription of the lac genes. Superrepressor mutations were dominant over lacI+; partial diploids with genotype lacIslacZ+/lacI+ lacZ+ were unable to synthesize either β-galactosidase or permease, whether or not lactose was present (Figure 16.11).
Operator Mutations
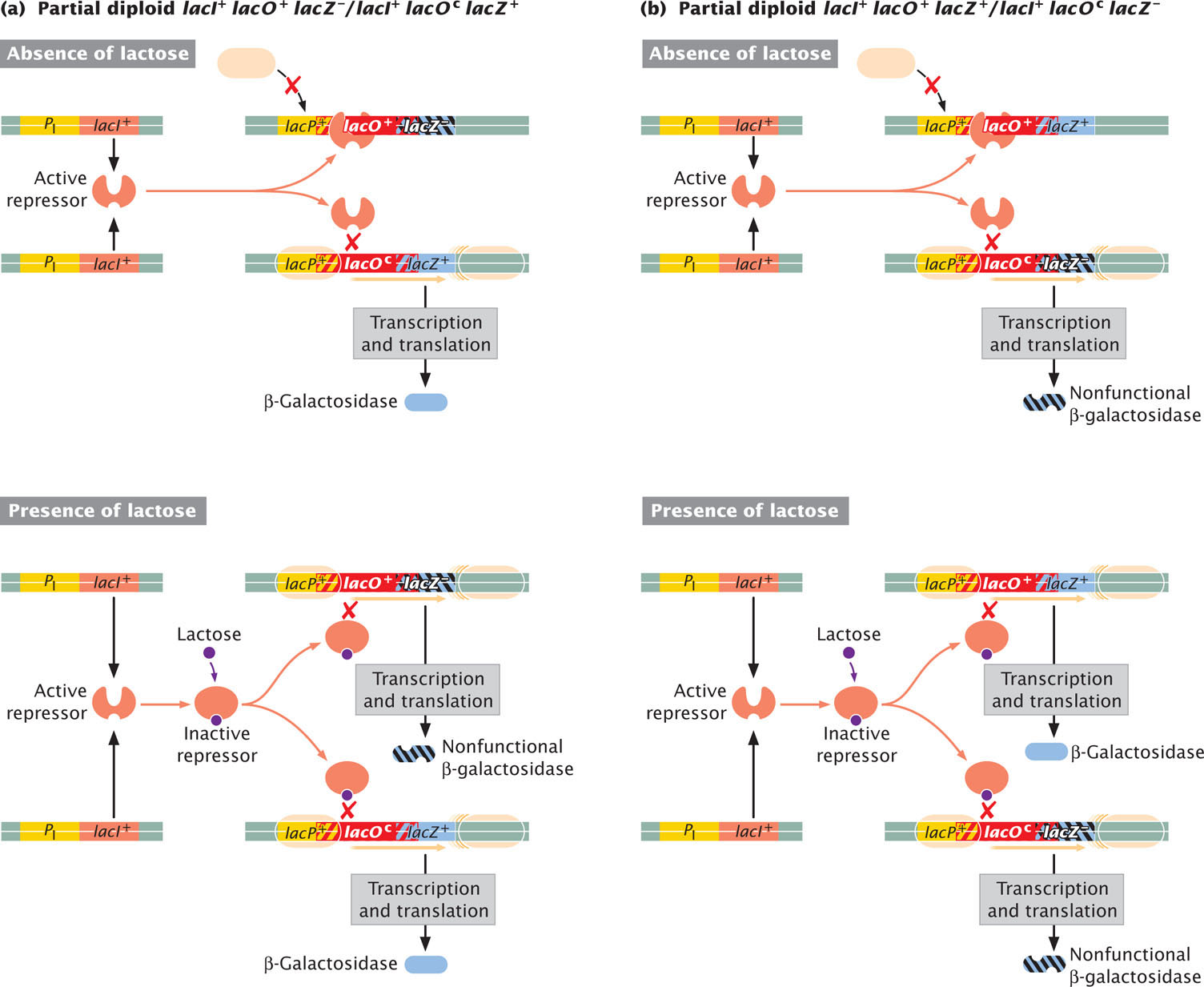
Jacob and Monod mapped another class of constitutive mutants to a site adjacent to lacZ. These mutations occurred at the operator site and were referred to as lacOc (O stands for operator and “c” for constitutive). The lacOc mutations altered the sequence of DNA at the operator so that the repressor protein was no longer able to bind. A partial diploid with genotype lacI+ lacOc lacZ+/lacI+ lacO+ lacZ+ exhibited constitutive synthesis of β-galactosidase, indicating that lacOc is dominant over lacO+.
Analysis of other partial diploids showed that the lacO gene is cis acting, affecting only genes on the same DNA molecule. For example, a partial diploid with genotype lacI+ lacO+ lacZ−/lacI+ lacOc lacZ+ was constitutive, producing β-galactosidase in the presence or absence of lactose(Figure 16.12a), but a partial diploid with genotype lacI+ lacO+ lacZ+/lacI+ lacOc lacZ− produced β-galactosidase only in the presence of lactose (Figure 16.12b). In the constitutive partial diploid (lacI+ lacO+ lacZ−/lacI+ lacOc lacZ+; see Figure 16.12a), the lacOc mutation and the functional lacZ+ gene are present on the same DNA molecule; but, in lacI+ lacO+ lacZ+/lacI+ lacOc lacZ− (see Figure 16.12b), the lacOc mutation and the functional lacZ+ gene are on different molecules. The lacO mutation affects only genes to which it is physically connected, as is true of all operator mutations. They prevent the binding of a repressor protein to the operator and thereby allow RNA polymerase to transcribe genes on the same DNA molecule. However, they cannot prevent a repressor from binding to normal operators on other DNA molecules. Watch Animation 16.1 to observe the effects of different-combinations of lacI and lacO mutations on the expression of the lac operon.
to observe the effects of different-combinations of lacI and lacO mutations on the expression of the lac operon.  TRY PROBLEM 21
TRY PROBLEM 21
Promoter Mutations
Mutations affecting lactose metabolism have also been isolated at the promoter site; these mutations are designated lacP−, and they interfere with the binding of RNA polymerase to the promoter. Because this binding is essential for the transcription of the structural genes, E. coli strains with lacP− mutations don’t produce lac proteins either in the presence or in the absence of lactose. Like operator mutations, lacP− mutations are cis acting and thus affect only genes on the same DNA molecule. The partial diploid lacI+ lacP+ lacZ+/lacI+lacP− lacZ+ exhibits normal synthesis of β-galactosidase, whereas lacI+ lacP− lacZ+/lacI+ lacP+ lacZ− fails to produce β-galactosi-dase whether or not lactose is present. The different types of mutations that occur in the lac operon are summarized in Table 16.2.
| Type | Location | Cis/Trans | Effect |
|---|---|---|---|
| Structural gene mutations | lacZ, lacY | Only affect lacZ or lacY | Alter amino acid sequence of protein encoded by gene in which mutation occurs |
| Regulator gene mutations | lacI | Trans | Affect transcription of structural genes |
| Operator mutations | lacO | Cis | Affect transcription of structural genes |
| Promoter mutations | lacP | Cis | Affect transcription of structural genes |
WORKED PROBLEM
For E. coli strains with the following lac genotypes, use a plus sign (+) to indicate the synthesis of β-galactosidase and permease and a minus sign (−) to indicate no synthesis of the proteins when lactose is absent and when it is present.
Genotype of strain
- a. lacI+ lacP+ lacO+ lacZ+ lacY+
- b. lacI+ lacP+ lacOc lacZ− lacY+
- c. lacI+ lacP− lacO+ lacZ+ lacY−
- d. lacI+ lacP+ lacO+ lacZ− lacY−/lacI− lacP+ lacO+ lacZ+ lacY+
What information is required in your answer to the problem?
An indication of whether or not β-galactosidase and permease are produced by each genotype when lactose is present and when lactose is absent, by placing a plus sign (+) or minus sign (−) for each enzyme and condition in the table.
What information is provided to solve the problem?
The genotype of each strain.
| Lactose absent | Lactose present | |||
|---|---|---|---|---|
| Genotype of strain | β-Galactosidase | Permease | β-Galactosidase | Permease |
| a. lacI+ lacP+ lacO+ lacZ+ lacY+ | − | − | + | + |
| b. lacI+ lacP+ lacOc lacZ− lacY+ | − | + | − | + |
| c. lacI+ lacP− lacO+ lacZ+ lacY− | − | − | − | − |
| d. lacI+ lacP+ lacO+ lacZ− lacY−/lacI− lacP+ lacO+ lacZ+ lacY+ | − | − | + | + |
- a. All the genes possess normal sequences so the lac operon functions normally: when lactose is absent, the regulator protein binds to the operator and inhibits the transcription of the structural genes, and so β-galactosidase and permease are not produced. When lactose is present, some of it is converted into allolactose, which binds to the repressor and makes it inactive; the repressor does not bind to the operator, and so the structural genes are transcribed and β-galactosidase and permease are produced.
- b. The structural lacZ gene is mutated so β-galactosidase will not be produced under any conditions. The lacO gene has a constitutive mutation, which means that the repressor is unable to bind to lacO, and so transcription takes place at all times. Therefore, permease will be produced in both the presence and the absence of lactose.
- c. In this strain, the promoter is mutated, and so RNA polymerase is unable to bind and transcription does not take place. Therefore, β-galactosidase and permease are not produced under any conditions.
- d. This strain is a partial diploid, which consists of two copies of the lac operon—one on the bacterial chromosome and the other on a plasmid. The lac operon represented in the upper part of the genotype has mutations in both the lacZ and the lacY genes, and so it is not capable of encoding β-galactosidase or permease under any conditions. The lac operon in the lower part of the genotype has a defective regulator gene, but the normal regulator gene in the upper operon produces a diffusible repressor (trans acting) that binds to the lower operon in the absence of lactose and inhibits transcription. Therefore, no β-galactosidase or permease is produced when lactose is absent. In the presence of lactose, the repressor cannot bind to the operator, and so the lower operon is transcribed and β-galactosidase and permease are produced.
Now try your own hand at predicting the outcome of different lac mutations by working Problem 19 at the end of the chapter.
Positive Control and Catabolite Repression
E. coli and many other bacteria metabolize glucose preferentially in the presence of lactose and other sugars. They do so because glucose enters glycolysis without further modification and therefore requires less energy to metabolize than do other sugars. When glucose is available, genes that participate in the metabolism of other sugars are repressed, in a phenomenon known as catabolite repression. For example, the efficient transcription of the lac operon takes place only if lactose is present and glucose is absent. But how is the expression of the lac operon influenced by glucose? What brings about catabolite repression?
Catabolite repression results from positive control in response to glucose. (This regulation is in addition to the negative control brought about by the repressor binding at the operator site of the lac operon when lactose is absent.) Positive control is accomplished through the binding of a dimeric protein called the catabolite activator protein (CAP) to a site that is about 22 nucleotides long and is located within or slightly upstream of the promoter of the lac genes (Figure 16.13). RNA polymerase does not bind efficiently to many promoters unless CAP is first bound to the DNA. Before CAP can bind to DNA, it must form a complex with a modified nucleotide called adenosine-3′, 5′-cyclic monophosphate (cyclic AMP, or cAMP), which is important in cellular signaling processes in both bacterial and eukaryotic cells. In E. coli, the concentration of cAMP is regulated so that its concentration is inversely proportional to the level of available glucose. A high concentration of glucose within the cell lowers the amount of cAMP so little cAMP-CAP complex is available to bind to the DNA. Consequently, RNA polymerase has poor affinity for the lac promoter, and little transcription of the lac operon takes place. Low concentrations of glucose stimulate high levels of cAMP, resulting in increased cAMP–CAP binding to DNA. This increase enhances the binding of RNA polymerase to the promoter and increases transcription of the lac genes by approximately 50-fold.
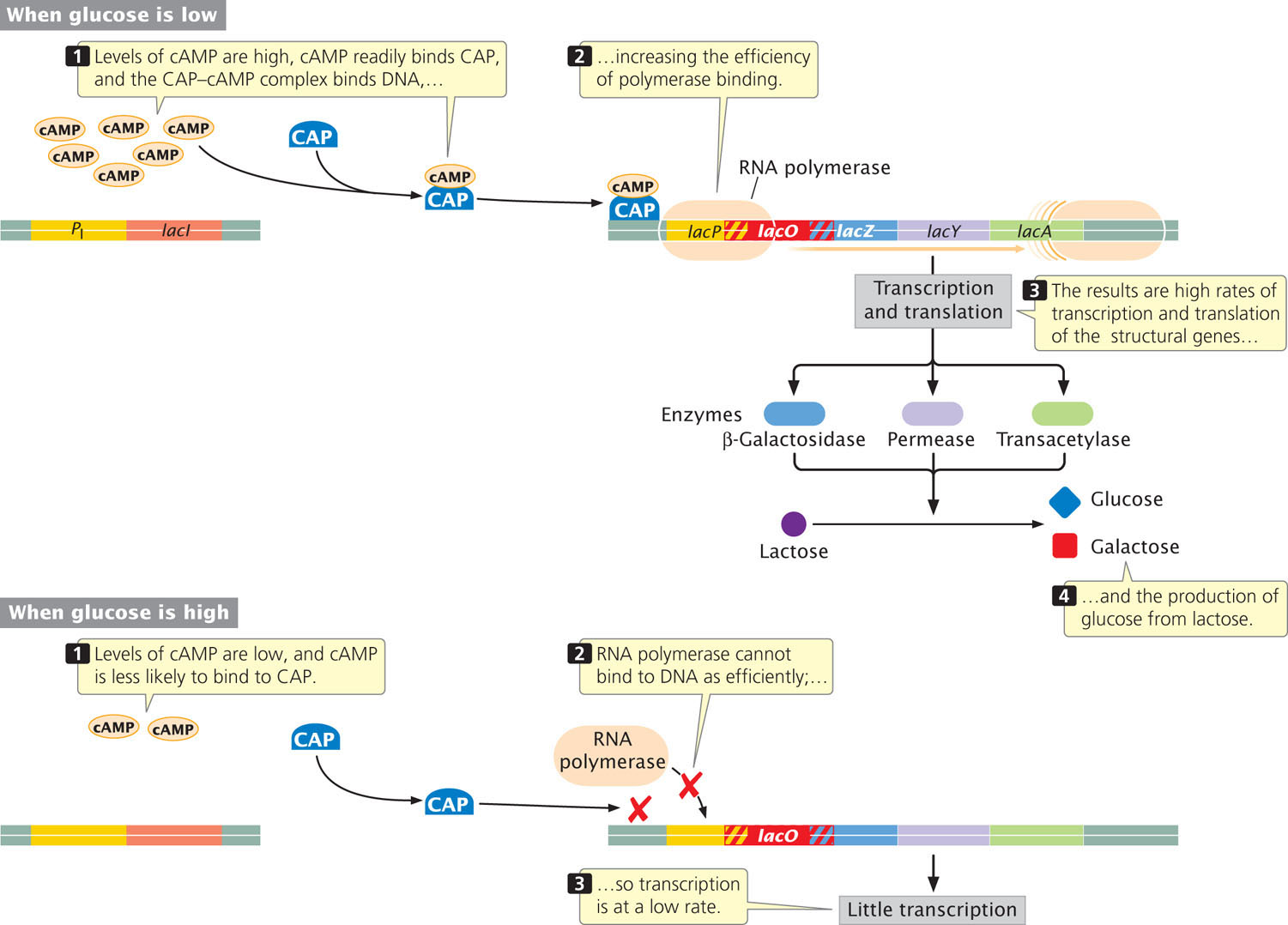
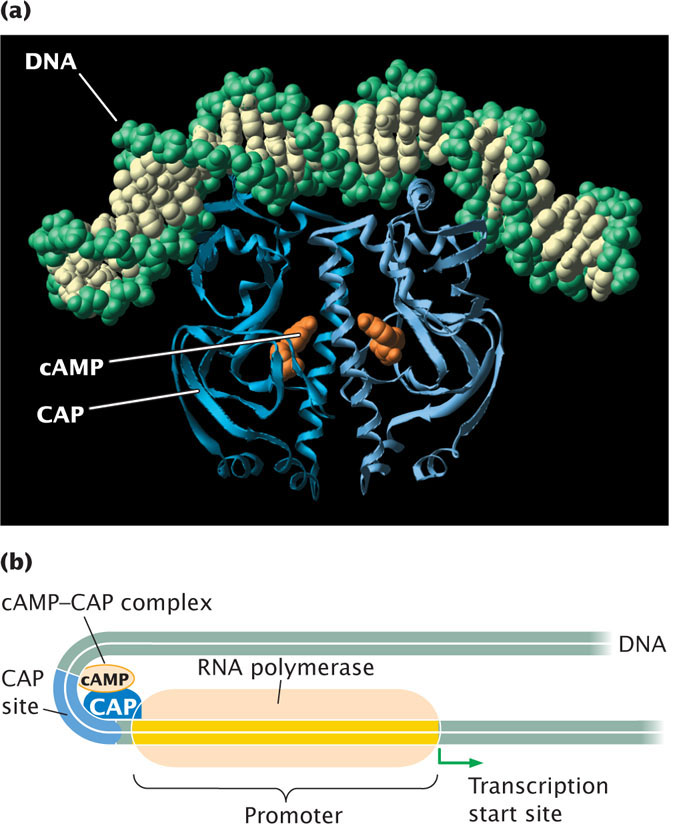
The catabolite activator protein exerts positive control in more than 20 operons of E. coli. The response to CAP varies among these promoters; some operons are activated by low levels of CAP, whereas others require high levels. CAP contains a helix-turn-helix DNA-binding motif and, when it binds at the CAP site on DNA, it causes the DNA helix to bend (Figure 16.14). The bent helix enables CAP to facilitate the binding of RNA polymerase at the promoter and the initiation of transcription.  TRY PROBLEM 14
TRY PROBLEM 14
CONCEPTS
In spite of its name, catabolite repression is a type of positive control in the lac operon. The catabolite activator protein (CAP), complexed with cAMP, binds to a site near the promoter and stimulates the binding of RNA polymerase. Cellular levels of cAMP are controlled by glucose; a low glucose level increases the abundance of cAMP and enhances the transcription of the lac structural genes.
 CONCEPT CHECK 7
CONCEPT CHECK 7What is the effect of high levels of glucose on the lac operon?
- Transcription is stimulated.
- Little transcription takes place.
- Transcription is not affected.
- Transcription may be stimulated or inhibited, depending on the levels of lactose.
The trp Operon of E. coli
The lac operon just discussed is an inducible operon, one in which transcription does not normally take place and must be turned on. Other operons are repressible; transcription in these operons is normally turned on and must be repressed. The tryptophan (trp) operon in E. coli, which controls the biosynthesis of the amino acid tryptophan, is an example of a negative repressible operon.
The trp operon contains five structural genes (trpE, trpD, trpC, trpB, and trpA) that produce the components of three enzymes (two of the enzymes consist of two polypeptide chains). These enzymes convert chorismate into tryptophan (Figure 16.15). The first structural gene, trpE, contains a long 5′ untranslated region (5′ UTR) that is transcribed but does not encode any of these enzymes. Instead, this 5′ UTR plays an important role in another regulatory mechanism, discussed in the next section. Upstream of the 5′ UTR is the trp promoter. When tryptophan levels are low, RNA polymerase binds to the promoter and transcribes the five structural genes into a single mRNA, which is then translated into enzymes that convert chorismate into tryptophan.
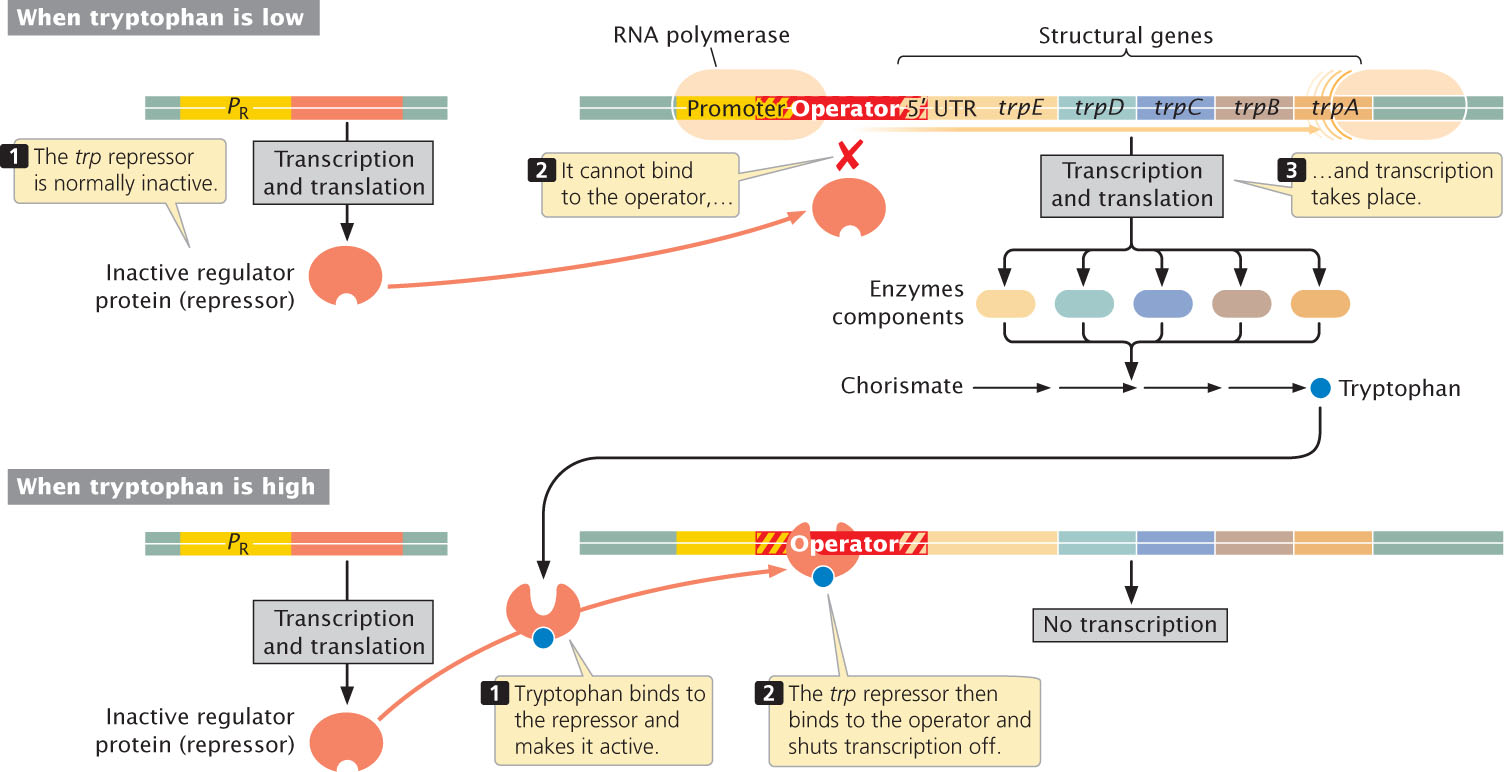
Some distance from the trp operon is a regulator gene, trpR, which encodes a repressor that alone cannot bind DNA (see Figure 16.15). Like the lac repressor, the tryptophan repressor has two binding sites, one that binds to DNA at the operator site and another that binds to tryptophan (the activator). Binding with tryptophan causes a conformational change in the repressor that makes it capable of binding to DNA at the operator site, which overlaps the promoter. When the operator is occupied by the tryptophan repressor, RNA polymerase cannot bind to the promoter and the structural genes cannot be transcribed. Thus, when cellular levels of tryptophan are low, transcription of the trp operon takes place and more tryptophan is synthesized; when cellular levels of tryptophan are high, transcription of the trp operon is inhibited and the synthesis of more tryptophan does not take place.
Bacterial Enhancers
Another type of regulatory sequence that affects transcription is an enhancer, a DNA element that affects transcription but, in contrast to promoters, is typically found some distance from the gene (see Chapter 17). Enhancers were originally described in eukaryotes, but research now indicates some also occur in bacteria and archaea.
Like enhancers in eukaryotes, bacterial enhancers contain binding sites for proteins that increase the rate of transcription from promoters that are distant from the gene. They do this by causing the DNA between the promoter and enhancer to loop out, so that the transcription factor at the enhancer directly interacts with RNA polymerase at the promoter. Enhancers are also position independent, meaning that they can be moved without affecting their ability to enhance transcription. Most bacterial enhancers are found upstream of genes that utilize a special type of sigma factor (see Chapter 13) known as sigma 54 (σ54). Enhancers will be discussed in more detail in Chapter 17.
CONCEPTS
The trp operon is a negative repressible operon that controls the biosynthesis of tryptophan. In a repressible operon, transcription is normally turned on and must be repressed: this is accomplished through the binding of tryptophan to the repressor, which renders the repressor active. The active repressor binds to the operator and prevents RNA polymerase from transcribing the structural genes. Bacterial enhancers increase the rate of transcription at genes that are distant from the enhancer.
 CONCEPT CHECK 8
CONCEPT CHECK 8In the trp operon, what happens to the trp repressor in the absence of tryptophan?
- It binds to the operator and represses transcription.
- It cannot bind to the operator and transcription takes place.
- It binds to the regulator gene and represses transcription.
- It cannot bind to the regulator gene and transcription takes place.