16.3 Some Operons Regulate Transcription Through Attenuation, the Premature Termination of Transcription
We’ve now seen several different ways in which a cell regulates the initiation of transcription in an operon. Some operons have an additional level of control that affects the continuation of transcription rather than its initiation. In attenuation, transcription begins at the start site, but termination takes place prematurely, before the RNA polymerase even reaches the structural genes. Attenuation takes place in a number of operons that encode enzymes participating in the biosynthesis of amino acids.
Attenuation in the trp Operon of E. coli
We can understand the process of attenuation most easily by looking at one of the best-studied examples, which is found in the trp operon of E. coli. The trp operon is unusual in that it is regulated both by repression and by attenuation. Most operons are regulated by one of these mechanisms but not by both of them.
Attenuation first came to light when Charles Yanofsky and his colleagues made several observations in the early 1970s that indicated that repression at the operator site is not the only method of regulation in the trp operon. They isolated a series of mutants that exhibited high levels of transcription, yet control at the operator site was unaffected, suggesting that some mechanism other than repression at the operator site was controlling transcription. Furthermore, they observed that two mRNAs of different sizes were transcribed from the trp operon: a long mRNA containing sequences for the structural genes and a much shorter mRNA of only 140 nucleotides. These observations led Yanofsky to propose that a mechanism that caused premature termination of transcription also regulates transcription in the trp operon.
Close examination of the trp operon reveals a region of 162 nucleotides that corresponds to the long 5′ UTR of the mRNA (mentioned earlier) transcribed from the trp operon (Figure 16.16a). The 5′ UTR (also called a leader) contains four regions: region 1 is complementary to region 2, region 2 is complementary to region 3, and region 3 is complementary to region 4 (Figure 16.16b). These complementarities allow the 5′ UTR to fold into two different secondary structures (Figure 16.16c). Only one of these secondary structures causes attenuation.
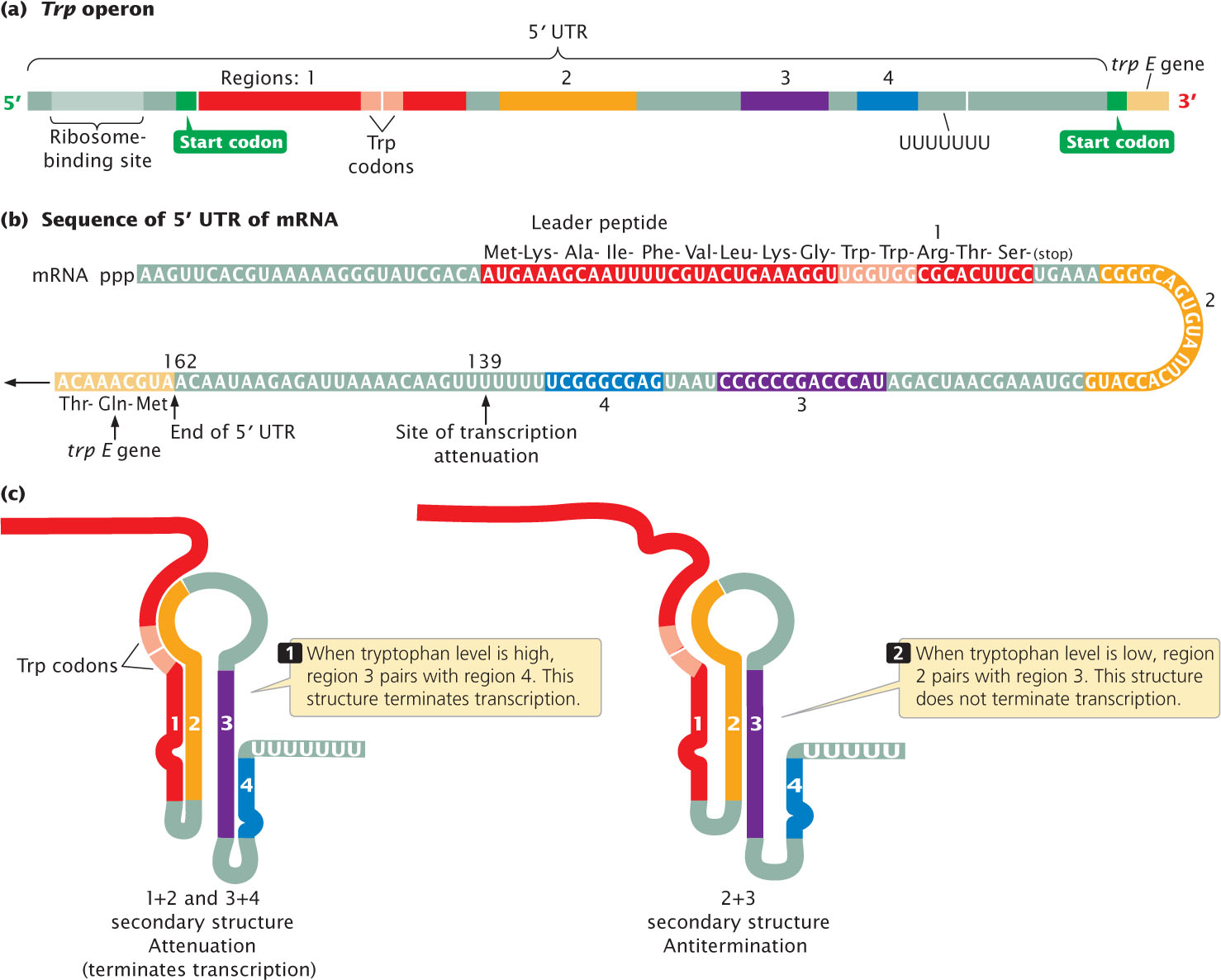
One of the secondary structures contains one hairpin produced by the base pairing of regions 1 and 2 and another hairpin produced by the base pairing of regions 3 and 4. Notice that a string of uracil nucleotides follows the 3+4 hairpin. Not coincidentally, the structure of a bacterial intrinsic terminator (see Chapter 13) includes a hairpin followed by a string of uracil nucleotides; this secondary structure in the 5′ UTR of the trp operon is indeed a terminator and is called an attenuator. The attenuator forms when cellular levels of tryptophan are high, causing transcription to be terminated before the trp structural genes can be transcribed.
When cellular levels of tryptophan are low, however, the alternative secondary structure of the 5′ UTR is produced by the base pairing of regions 2 and 3 (see Figure 16.16b). This base pairing also produces a hairpin, but this hairpin is not followed by a string of uracil nucleotides; so this structure does not function as a terminator. RNA polymerase continues past the 5′ UTR into the coding section of the structural genes, and the enzymes that synthesize tryptophan are produced. Because it prevents the termination of transcription, the 2+3 structure is called an antiterminator.
To summarize. the 5′ UTR of the trp operon can fold into one of two structures. When the tryptophan level is high, the 3+4 structure forms, transcription is terminated within the 5′ UTR, and no additional tryptophan is synthesized. When the tryptophan level is low, the 2+3 structure forms, transcription continues through the structural genes, and tryptophan is synthesized. The critical question, then, is: Why does the 3+4 structure arise when the level of tryptophan in the cell is high, whereas the 2+3 structure arises when the level is low?
To answer this question, we must take a closer look at the nucleotide sequence of the 5′ UTR. At the 5′ end, upstream of region 1, is a ribosome-binding site (see Figure 16.16a). Region 1 encodes a small protein. Within the coding sequence for this protein are two UGG codons, which Specify the amino acid tryptophan; so tryptophan is required for the translation of this 5′ UTR sequence. The small protein encoded by the 5′ UTR has not been isolated and is presumed to be unstable; its only apparent function is to control attenuation. Although it was stated in Chapter 14 that a 5′ UTR is not translated into a protein, the 5′ UTR of operons subject to attenuation is an exception to this rule. The precise timing and interaction of transcription and translation in the 5′ UTR determine whether attenuation takes place.
Transcription when Tryptophan Levels are Low
Let’s first consider what happens when intracellular levels of tryptophan are low. Recall that, in prokaryotic cells, transcription and translation are coupled: while transcription is taking place at the 3′ end of the mRNA, translation is initiated at the 5′ end. RNA polymerase begins transcribing the DNA, producing region 1 of the 5′ UTR (Figure 16.17a). Closely following RNA polymerase, a ribosome binds to the 5′ UTR and begins to translate the coding region. Meanwhile, RNA polymerase is transcribing region 2 (Figure 16.17b). Region 2 is complementary to region 1 but, because the ribosome is translating region 1, the nucleotides in regions 1 and 2 cannot base pair.
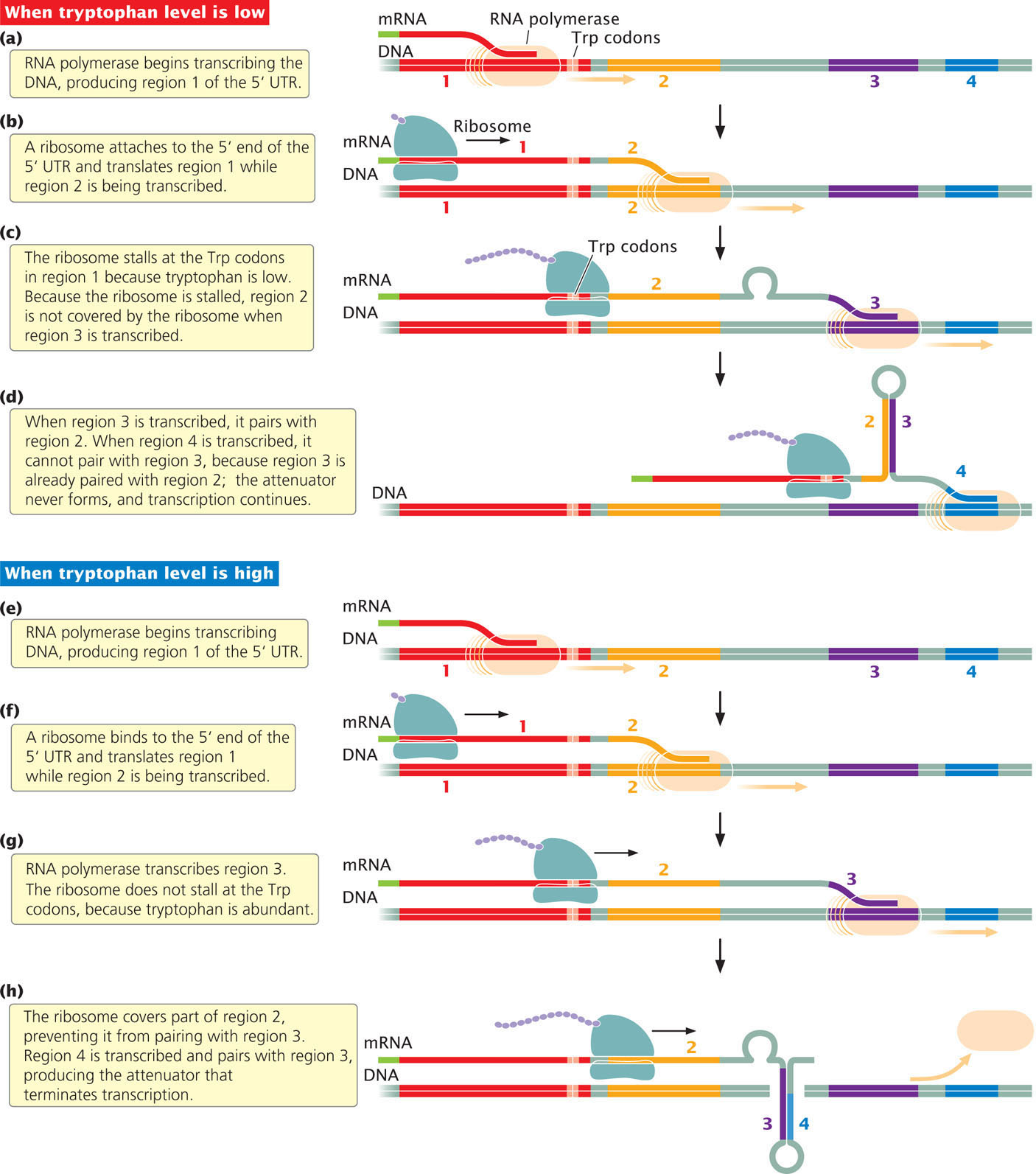
RNA polymerase begins to transcribe region 3, and the ribosome reaches the UGG tryptophan codons in region 1. When it reaches the tryptophan codons, the ribosome stalls (Figure 16.17c) because the level of tryptophan is low and tRNAs charged with tryptophan are scarce or even unavailable. The ribosome sits at the tryptophan codons, awaiting the arrival of a tRNA charged with tryptophan. Stalling of the ribosome does not, however, hinder transcription; RNA polymerase continues to move along the DNA, and transcription gets ahead of translation.
Because the ribosome is stalled at the tryptophan codons in region 1, region 2 is free to base pair with region 3, forming the 2+3 hairpin (Figure 16.17d). This hairpin does not cause termination, and so transcription continues. Because region 3 is already paired with region 2, the 3+4 hairpin (the attenuator) never forms, and so attenuation does not take place and transcription continues. RNA polymerase continues along the DNA, past the 5′ UTR, transcribing all the structural genes into mRNA, which is translated into the enzymes encoded by the trp operon. These enzymes then synthesize more tryptophan.
Transcription when Tryptophan Levels are High
Now let’s see what happens when intracellular levels of tryptophan are high. Once again, RNA polymerase begins transcribing the DNA, producing region 1 of the 5′ UTR (Figure 16.17e). Closely following RNA polymerase, a ribosome binds to the 5′ UTR and begins to translate the coding region (Figure 16.17f). When the ribosome reaches the two UGG tryptophan codons, it doesn’t slow or stall, because tryptophan is abundant and tRNAs charged with tryptophan are readily available (Figure 16.17g). This point is critical to note: because tryptophan is abundant, translation can keep up with transcription.
As it moves past region 1, the ribosome partly covers region 2 (Figure 16.17h); meanwhile, RNA polymerase completes the transcription of region 3. Although regions 2 and 3 are complementary, the ribosome physically blocks their pairing.
RNA polymerase continues to move along the DNA, eventually transcribing region 4 of the 5′ UTR. Region 4 is complementary to region 3, and, because region 3 cannot base pair with region 2, it pairs with region 4. The pairing of regions 3 and 4 (see Figure 16.17h) produces the attenuator and transcription terminates just beyond region 4. The structural genes are not transcribed, no tryptophan-producing enzymes are translated, and no additional tryptophan is synthesized. Important events in the process of attenuation are summarized in Table 16.3. Try pausing the ribosome at the trp codon for different lengths of time in  Animation 16.2 and see what effect the pause has on transcription.
Animation 16.2 and see what effect the pause has on transcription.
| Intracellular Level of Tryptophan | Ribosome Stalls at Trp Codons | Position of Ribosome When Region 3 Is Transcribed | Secondary Structure of 5′ UTR | Termination of Transcription of trp Operon |
|---|---|---|---|---|
| High | No | Covers region 2 | 3+4 hairpin | Yes |
| Low | Yes | Covers region 1 | 2 + 3 hairpin | No |
A key factor controlling attenuation is the number of tRNA molecules charged with tryptophan, because their availability is what determines whether the ribosome stalls at the tryptophan codons. A second factor concerns the synchronization of transcription and translation, which is critical to attenuation. Synchronization is achieved through a pause site located in region 1 of the 5′ UTR. When this site is transcribed, the RNA folds into a secondary structure that inhibits further transcription. Thus, the RNA polymerase stops temporarily at the pause site, allowing time for a ribosome to bind to the 5′ end of the mRNA. As the ribosome approaches the secondary structure in the RNA, the ribosome disrupts it and allows transcription to continue. Translation then closely follows transcription. It is important to point out that ribosomes do not traverse the convoluted hairpins of the 5′ UTR to translate the structural genes. Ribosomes that attach to the 5′ end of region 1 of the mRNA encounter a stop codon at the end of region 1. New ribosomes translating the structural genes attach to a different ribosome-binding site located near the beginning of the trpE gene.  TRY PROBLEM 27
TRY PROBLEM 27
Why Does Attenuation Take Place in the trp Operon?
Why do bacteria need attenuation in the trp operon? Shouldn’t repression at the operator site prevent transcription from taking place when tryptophan levels in the cell are high? Why does the cell have two types of control? Part of the answer is that repression is never complete; some transcription is initiated even when the trp repressor is active; repression reduces transcription only as much as 70-fold. Attenuation can further reduce transcription another 8- to 10-fold, so together the two processes are capable of reducing transcription of the trp operon more than 600-fold. Both mechanisms provide E. coli with a much finer degree of control over tryptophan synthesis than either could achieve alone.
Another reason for the dual control is that attenuation and repression respond to different signals: repression responds to the cellular levels of tryptophan, whereas attenuation responds to the number of tRNAs charged with tryptophan. There may be times when a cell’s ability to respond to these different signals is advantageous. Finally, the trp repressor affects several operons other than the trp operon. At an earlier stage in the evolution of E. coli, the trp operon may have been controlled only by attenuation. The trp repressor may have evolved primarily to control the other operons and only incidentally affects the trp operon.
Attenuation is a difficult process to grasp because you must simultaneously visualize how two dynamic processes—transcription and translation—interact, and it’s easy to confuse them. Remember that attenuation refers to the early termination of transcription, not translation (although events in translation bring about the termination of transcription). Attenuation often causes confusion because we know that transcription must precede translation. We’re comfortable with the idea that transcription might affect translation, but it’s harder to imagine that the effects of translation could influence transcription, as they do in attenuation. The reality is that transcription and translation are closely coupled in prokaryotic cells, and events in one process can easily affect the other.
CONCEPTS
In attenuation, transcription is initiated but terminates prematurely. When tryptophan levels are low, the ribosome stalls at the tryptophan codons and transcription continues. When tryptophan levels are high, the ribosome does not stall at the tryptophan codons, and the 5′ UTR adopts a secondary structure that terminates transcription before the structural genes can be copied into RNA (attenuation).
 CONCEPT CHECK 9
CONCEPT CHECK 9Attenuation results when which regions of the 5′ UTR pair?
- 1 and 3
- 2 and 3
- 2 and 4
- 3 and 4