Termination
Protein synthesis ends when the ribosome translocates to a termination codon. Because there are no tRNAs with anticodons complementary to the termination codons, no tRNA enters the A site of the ribosome (Figure 11.12a). Instead, proteins called release factors bind to the ribosome (Figure 11.12b). Escherichia coli has three release factors: RF-
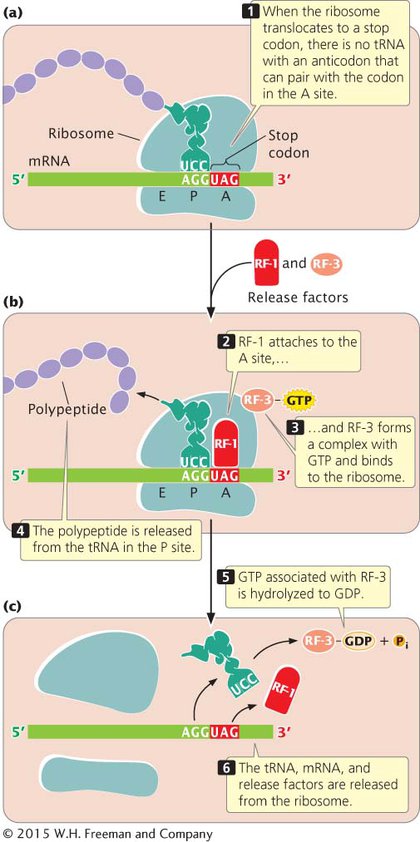
It is important to note that the termination codon is not located at the 3′ end of the mRNA; rather, the termination codon is followed by a number of nucleotides that constitute the 3′ untranslated region (UTR) of the mRNA. The 3′ UTR often contains sequences that affect the stability of the mRNA and influence whether translation takes place (see Chapter 12).
Recent research shows that some bacterial ribosomes engage in a type of proofreading, similar to the proofreading that DNA polymerases perform during replication. After translocation, the ribosome checks the interaction between the mRNA and the tRNA in the P site. If the wrong tRNA was added, the alignment between the mRNA and tRNA will be incorrect, which triggers premature termination of translation. Evidence suggests that an important function of RF-
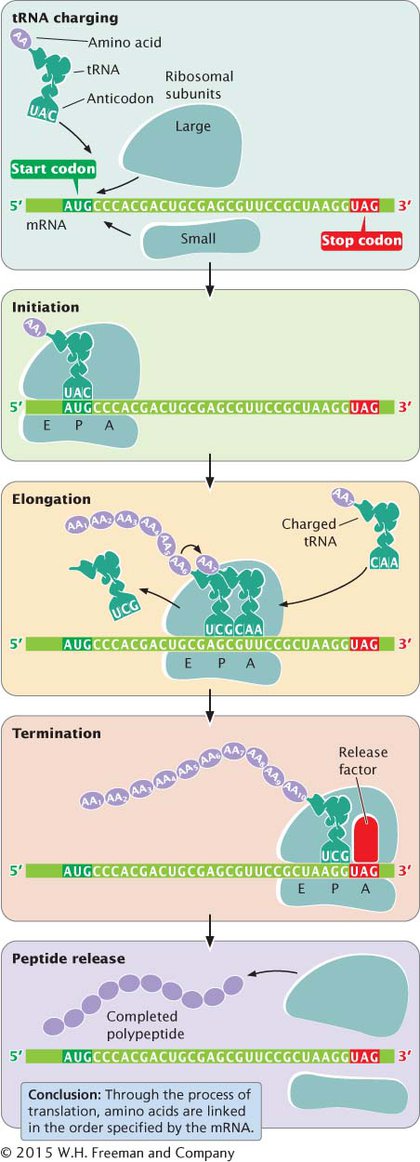
The overall process of protein synthesis, including tRNA charging, initiation, elongation, and termination, is summarized in Figure 11.13. The components taking part in this process are listed in Table 11.2.
| Stage | Component | Function |
|---|---|---|
| tRNA charging |
Amino acids tRNAs Aminoacyl- ATP |
Building blocks of proteins Deliver amino acids to ribosomes Attach amino acids to tRNAs Provides energy for binding amino acids to tRNAs |
| Initiation |
mRNA fMet- 30S ribosomal subunit 50S ribosomal subunit Initiation factor 1 Initiation factor 2 Initiation factor 3 |
Carries coding instructions Provides first amino acid in peptide Attaches to mRNA Stabilizes tRNAs and amino acids Enhances dissociation of large and small subunits of ribosome Binds GTP; delivers fMet- Binds to 30S subunit and prevents association with 50S subunit |
| Elongation |
70S initiation complex Charged tRNAs Elongation factor Tu Elongation factor Ts Elongation factor G GTP rRNA in 50S ribosomal subunit |
Functional ribosome with A, P, and E sites where protein synthesis takes place Bring amino acids to ribosome and help assemble them in order specified by mRNA Binds GTP and charged tRNA; delivers charged tRNA to A site Regenerates active elongation factor Tu Stimulates movement of ribosome to next codon Provides energy Creates peptide bond between amino acids in A site and P site |
| Termination | Release factors 1, 2, and 3 | Bind to ribosome when stop codon is reached and terminate translation |
CONCEPTS
Translation ends when the ribosome reaches a termination codon. Release factors bind to the termination codon, causing the release of the polypeptide from the last tRNA, of the tRNA from the ribosome, and of the mRNA from the ribosome.
CONNECTING CONCEPTS
A Comparison of Bacterial and Eukaryotic Translation
We have now considered the process of translation in bacterial cells and noted some distinctive differences that exist in eukaryotic cells. Let’s reflect on some of the important similarities and differences between protein synthesis in bacterial and in eukaryotic cells.
First, we should emphasize that the genetic code of bacterial and eukaryotic cells is virtually identical; the only difference is in the amino acid specified by the initiation codon. In bacterial cells, AUG encodes a modified type of methionine, N-formylmethionine, whereas in eukaryotic cells, AUG encodes unformylated methionine. One consequence of the fact that bacteria and eukaryotes use the same code is that eukaryotic genes can be translated in bacterial systems, and vice versa; this feature makes genetic engineering possible, as we will see in Chapter 14.
Another difference is that transcription and translation take place simultaneously in bacterial cells, but the nuclear envelope separates these processes in eukaryotic cells. The physical separation of transcription and translation has important implications for the control of gene expression, which we will consider in Chapter 12, and it allows for extensive modification of eukaryotic mRNAs, as discussed in Chapter 10.
Yet another difference is that mRNA in bacterial cells is short-
In both bacterial and eukaryotic cells, aminoacyl-
Other fundamental differences lie in the process of initiation. In bacterial cells, the small subunit of the ribosome attaches directly to the region surrounding the start codon through hydrogen bonding between the Shine–Dalgarno consensus sequence in the 5′ untranslated region of the mRNA and a sequence at the 3′ end of the 16S rRNA. In contrast, the small subunit of a eukaryotic ribosome first binds to proteins attached to the 5′ cap on mRNA and then migrates down the mRNA, scanning the sequence until it encounters the first AUG initiation codon. Additionally, a larger number of initiation factors take part in eukaryotic initiation than in bacterial initiation.
Elongation and termination are similar in bacterial and eukaryotic cells, although different elongation and termination factors are used. In both types of organisms, mRNAs are translated multiple times and are simultaneously attached to several ribosomes, as we will see next.