Gene Expression and Microarrays
Many important clues about gene function come from knowing when and where the genes are expressed. The development of microarrays has allowed the expression of thousands of genes to be monitored simultaneously.
Microarrays rely on nucleic acid hybridization, in which a known DNA fragment is used as a probe to find complementary sequences (Figure 15.7). Numerous known DNA fragments are fixed to a solid support in an orderly pattern or array, usually as a series of dots. These DNA fragments (the probes) usually correspond to known genes from a particular organism. An array containing tens of thousands of probes can be applied to a glass slide or silicon chip just a few square centimeters in size.
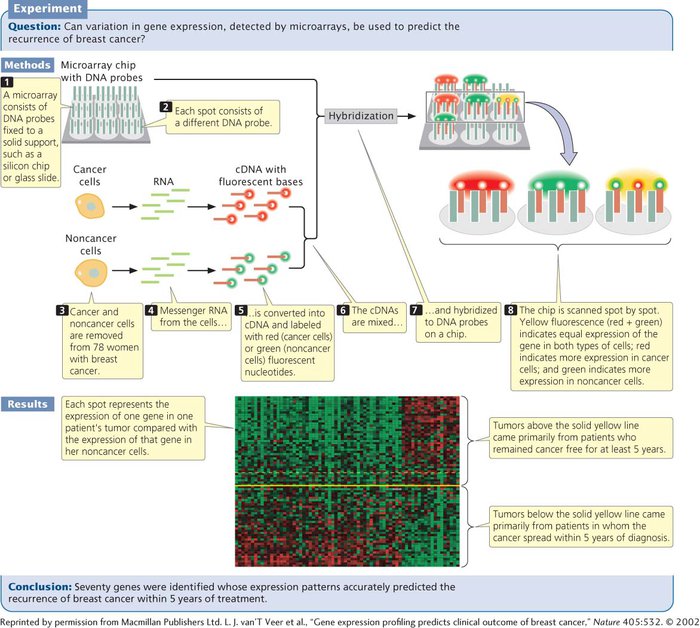
After the microarray has been constructed, mRNA, DNA, or cDNA isolated from experimental cells is labeled with fluorescent nucleotides and applied to the array. Any of the DNA or RNA molecules that are complementary to probes on the array will hybridize with them and emit fluorescence, which can be detected by an automated scanner.
Used with cDNA, microarrays can provide information about the expression of thousands of genes, enabling scientists to determine which genes are active in particular tissues. They can also be used to investigate how gene expression changes in the course of biological processes such as development or disease progression. In one study, researchers used microarrays to examine the expression patterns of 25,000 genes from primary tumors of 78 young women who had breast cancer (see Figure 15.7). Messenger RNA from cancer and noncancer cells was converted into cDNA and labeled with red and green fluorescent nucleotides, respectively. The labeled cDNAs were mixed and hybridized to a DNA chip, which contained DNA probes from numerous genes. Hybridization of the red (cancer) and green (noncancer) cDNA was proportional to the relative amounts of mRNA in the samples. The fluorescence of each spot was assessed by microscopic scanning and appeared as a single color. Red indicated the overexpression of a gene in the cancer cells relative to that in the noncancer cells (more red-
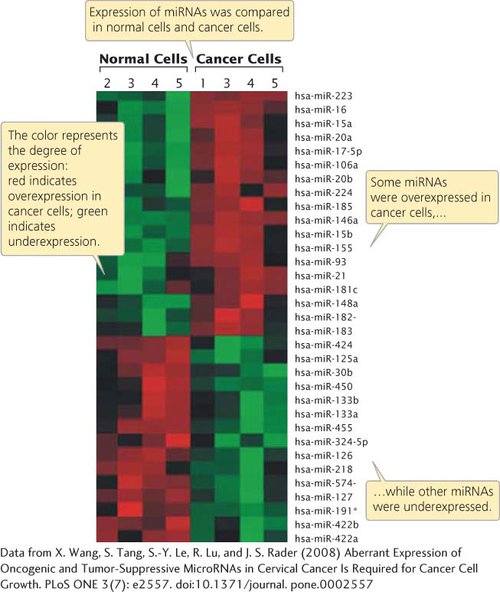
Researchers have also used microarrays to examine the expression of microRNAs (miRNAs) in human cancers. Recent research indicates that miRNAs are frequently expressed abnormally in cancerous tissue and may contribute to the progression of cancer (see Chapter 16). For example, one study using microarrays found that several miRNAs were overexpressed in cancerous cervical tissue compared with normal cervical tissue, while other miRNAs were underexpressed (Figure 15.8). Other studies using microarrays have demonstrated that miRNA expression is associated with resistance of tumors to chemotherapy and radiation, and that miRNA expression can be used to predict the responses of some tumors to cancer treatment. Results such as these suggest that gene- TRY PROBLEM 18
TRY PROBLEM 18
CONCEPTS
Microarrays, consisting of DNA probes attached to a solid support, can be used to determine which RNA molecules are being synthesized and can thus be used to examine changes in gene expression.