APPLICATION QUESTIONS AND PROBLEMS
Question 13
13.A 22-
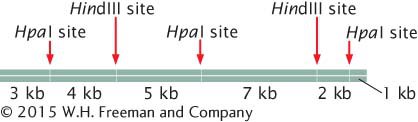
A batch of this DNA is first fully digested by HpaI alone, then another batch is fully digested by HindIII alone, and, finally, a third batch is fully digested by both HpaI and HindIII together. The fragments resulting from each of the three digestions are placed in separate wells of an agarose gel, separated by gel electrophoresis, and stained by ethidium bromide. Draw the bands as they would appear on the gel.
Question 14
*14.A linear piece of DNA that is 14 kb long is cut first by EcoRI alone, then by SmaI alone, and, finally, by both EcoRI and SmaI together. The following results are obtained:
| Digestion by EcoRI alone | Digestion by SmaI alone | Digestion by both EcoRI and SmaI |
|---|---|---|
| 3- |
7- |
2- |
| 5- |
7- |
3- |
| 6- |
4- |
|
| 5- |
Draw a map of the EcoRI and SmaI restriction sites on this 14-
Question 15
*15.A linear piece of DNA was broken into random, overlapping fragments and each fragment was sequenced. The sequence of each fragment is shown below.
Fragment 1: 5′—TAGTTAAAAC—
3′ Fragment 2: 5′—ACCGCAATACCCTAGTTAAA—
3′ Fragment 3: 5′—CCCTAGTTAAAAC—
3′ Fragment 4: 5′—ACCGCAATACCCTAGTT—
3′ Fragment 5: 5′—ACCGCAATACCCTAGTTAAA—
3′ Fragment 6: 5′—ATTTACCGCAAT—
3′
On the basis of overlap in sequence, assemble the fragments into a contig.
Question 16
16.In recent years, honeybee colonies throughout North America have been decimated by colony collapse disorder (CCD), which results in the rapid death of worker bees. First noticed by beekeepers in 2004, the disorder has been responsible for the loss of 50% to 90% of beekeeping operations in the United States. Evidence suggests that CCD is caused by a pathogen. Diana Cox-
| Potential pathogen | CCD hives infected (n = 30) |
Non- (n = 21) |
|---|---|---|
| Israeli acute paralysis virus | 83.3% | 4.8% |
| Kashmir bee virus | 100% | 76.2% |
| Nosema apis | 90% | 47.6% |
| Nosema cernae | 100% | 80.8% |
Question 17
17.James Noonan and his colleagues (J. Noonan et al. 2005. Science 309:597–599) set out to study the genome sequence of an extinct species of cave bear. They extracted DNA from 40,000-

Question 18
*18.Microarrays can be used to determine relative levels of gene expression. In one type of microarray, hybridization of red (experimental) and green (control) cDNAs is proportional to the relative amounts of mRNA in the samples. Red indicates the overexpression of a gene and green indicates the underexpression of a gene in the experimental cells relative to the control cells, yellow indicates equal expression in experimental and control cells, and no color indicates no expression in either experimental or control cells.

In one experiment, mRNA from a strain of antibiotic-
Question 19
19.Of the genes illustrated in the microarray shown in the lower part of Figure 15.7, are most overexpressed or underexpressed in tumors from patients that remained cancer free for at least five years? Explain your reasoning.
Question 20
20.Dictyostelium discoideum is a soil-
| Feature | D. discoideum | P. falciparum | S. cerevisiae | A. thaliana | D. melanogaster | C. elegans | H. sapiens |
|---|---|---|---|---|---|---|---|
| Organism | Amoeba | Malaria parasite | Yeast | Plant | Fruit fly | Nematode | Human |
| Cellularity | ? | Uni | Uni | Multi | Multi | Multi | Multi |
| Genome size (millions of base pairs) | 34 | 23 | 13 | 125 | 180 | 103 | 2,851 |
| Number of genes | 12,500 | 5,268 | 5,538 | 25,498 | 13,676 | 19,893 | 22,287 |
| Average gene length (bp) | 1,756 | 2,534 | 1,428 | 2,036 | 1,997 | 2,991 | 27,000 |
| Genes with introns (%) | 69 | 54 | 5 | 79 | 38 | 5 | 85 |
| Mean number of introns | 1.9 | 2.6 | 1.0 | 5.4 | 4.0 | 5.0 | 8.1 |
| Mean intron size (bp) | 146 | 179 | nd* | 170 | nd* | 270 | 3,365 |
| Mean G + C (exons) | 27% | 24% | 28% | 28% | 55% | 42% | 45% |
*nd = Not determined.
On the basis of the organisms other than D. discoideum listed in the table, what are some differences in genome characteristics between unicellular and multicellular organisms?
 Dictyostelium discoideum.[David Scharf/Science Source.]
Dictyostelium discoideum.[David Scharf/Science Source.]On the basis of these data, do you think that the genome of D. discoideum is more like those of unicellular eukaryotes or more like those of multicellular eukaryotes? Explain your answer.
Question 21
*21.A group of 250 scientists sequenced and analyzed the genomes of 12 species of Drosophila (Drosophila 12 Genomes Consortium. 2007. Nature 450:203–218). Data on genome sizes and numbers of protein-
Question 22
*22.A scientist determines the complete genomes and proteomes of a liver cell and a muscle cell from the same person. Would you expect bigger differences in the genomes or in the proteomes of these two cell types? Explain your answer.