Eukaryotic Chromosomes
Individual eukaryotic chromosomes contain enormous amounts of DNA. Like a bacterial chromosome, each eukaryotic chromosome consists of a single, extremely long linear molecule of DNA. Fitting all of this DNA into the nucleus requires tremendous packing and folding, the extent of which must change in the course of the cell cycle. The chromosomes are in an elongated, relatively uncondensed state during interphase (Chapter 2), but the term relatively is important here. Although the DNA of interphase chromosomes is less tightly packed than the DNA of mitotic chromosomes, it is still highly condensed; it’s just less condensed. In the course of the cell cycle, the level of DNA packing changes: chromosomes progress from a highly packed state to a state of extreme condensation, which is necessary for chromosome movement in mitosis and meiosis. DNA packing also changes locally during replication and transcription, when the two nucleotide strands must unwind so that particular base sequences are exposed. Thus, the packing of eukaryotic DNA (its tertiary chromosomal structure) is not static; rather, it changes regularly in response to cellular processes.
CHROMATIN STRUCTURE Eukaryotic DNA in the cell is closely associated with proteins. This combination of DNA and protein is called chromatin. The two basic types of chromatin are euchromatin, which undergoes the normal process of condensation and decondensation in the cell cycle, and heterochromatin, which remains in a highly condensed state throughout the cell cycle, even during interphase. Euchromatin constitutes the majority of the chromosomal material and is where most transcription takes place. All chromosomes have constitutive (permanent) heterochromatin at the centromeres and telomeres; the Y chromosome also consists largely of constitutive heterochromatin. Heterochromatin may also occur during certain developmental stages; this material is referred to as facultative heterochromatin. For example, facultative heterochromatin occurs along one entire X chromosome in female mammals when that X becomes inactivated (Chapter 4). In addition to remaining condensed throughout the cell cycle, heterochromatin is characterized by a general lack of transcription, the absence of crossing over, and replication late in S phase. Differences between euchromatin and heterochromatin are summarized in Table 8.3.
| Characteristic | Euchromatin | Heterochromatin |
|---|---|---|
| Chromatin condensation | Less condensed | More condensed |
| Location | On chromosome arms | At centromeres, telomeres, and other specific places |
| Type of sequences | Unique sequences | Repeated sequences* |
| Presence of genes | Many genes | Few genes* |
| When replicated | Throughout S phase | Late S phase |
| Transcription | Often | Infrequent |
| Crossing over | Common | Uncommon |
|
*Applies only to constitutive heterochromatin. |
||
The most abundant proteins in chromatin are the histones, which are small, positively charged proteins of five major types: H1, H2A, H2B, H3, and H4. All histones have a high percentage of arginine and lysine, positively charged amino acids that give the histones a net positive charge. Their positive charges attract the negative charges on the phosphates of DNA; this attraction holds the DNA in contact with the histones. A heterogeneous assortment of nonhistone chromosomal proteins is also found in eukaryotic chromosomes. At times, variant histones, with somewhat different amino acid sequences, are incorporated into chromatin in place of one of the major histone types.  TRY PROBLEM 31
TRY PROBLEM 31
CONCEPTS
Chromatin, which consists of DNA closely associated with proteins, is the material that makes up eukaryotic chromosomes. The most abundant of these proteins are the five types of positively charged histone proteins: H1, H2A, H2B, H3, and H4. Variant histones may at times be incorporated into chromatin in place of the normal histones.
 CONCEPT CHECK 9
CONCEPT CHECK 9
Neutralizing their positive charges would have which effect on the histone proteins?
They would bind the DNA tighter.
They would bind less tightly to the DNA.
They would no longer be attracted to each other.
They would cause supercoiling of the DNA.
b
THE NUCLEOSOME Chromatin has a highly complex structure with several levels of organization (Figure 8.18). The simplest level is the double-
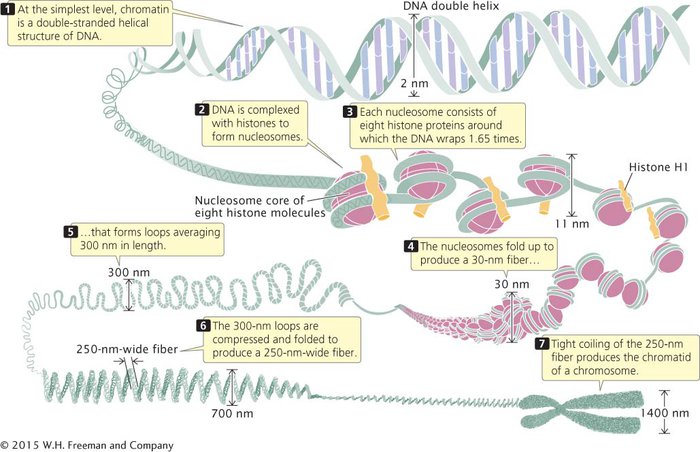
When chromatin is isolated from the nucleus of a cell and viewed with an electron microscope, it frequently looks like beads on a string (Figure 8.19a). If a small amount of nuclease is added to this structure, the enzyme cleaves the “string” between the “beads,” leaving individual beads attached to about 200 bp of DNA (Figure 8.19b). If more nuclease is added, the enzyme chews up all of the DNA between the beads and leaves a core of proteins attached to a fragment of DNA (Figure 8.19c). Such experiments demonstrated that chromatin is not a random association of proteins and DNA; rather, it has a fundamental repeating structure.
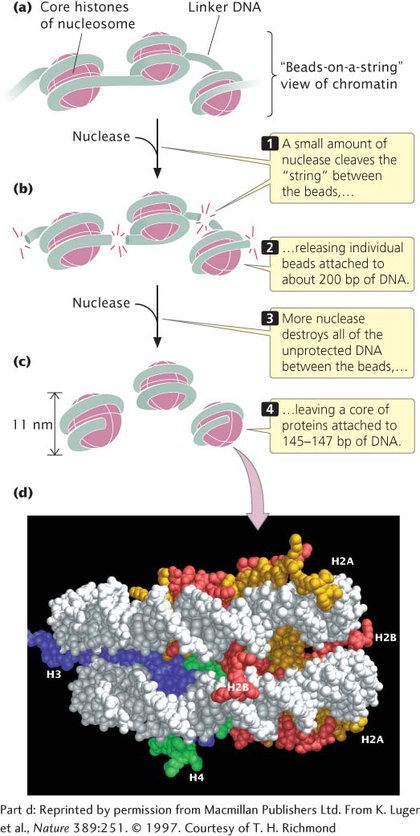
The repeating core of protein and DNA produced by digestion with nuclease enzymes is the simplest level of chromatin structure, the nucleosome (see Figure 8.18). The nucleosome is a core particle consisting of DNA wrapped about two times around an octamer of eight histone proteins (two copies each of H2A, H2B, H3, and H4), much like thread wound around a spool (Figure 8.19d). The DNA in direct contact with the histone octamer is between 145 and 147 bp in length.
Each of the histone proteins that make up the nucleosome core particle has a flexible “tail,” containing from 11 to 37 amino acids, which extends out from the nucleosome. Positively charged amino acids in the tails of the histones interact with the negative charges of the phosphates on the DNA, keeping the DNA and histones tightly associated (complexed). The tails of one nucleosome may also interact with neighboring nucleosomes, which facilitates compaction of the nucleosomes themselves. Chemical modifications of these histone tails bring about changes in chromatin structure that are necessary for gene expression.
The fifth type of histone, H1, is not a part of the nucleosome core particle, but plays an important role in nucleosome structure. H1 binds to 20–22 bp of DNA where the DNA joins and leaves the histone octamer (see Figure 8.18). It helps to lock the DNA into place, acting as a clamp around the nucleosome.
Each nucleosome encompasses about 167 bp of DNA. Nucleosomes are located at regular intervals along the DNA molecule and are separated from one another by linker DNA, which varies in size among cell types; in most cells, linker DNA comprises from about 30 to 40 bp. Nonhistone chromosomal proteins may be associated with this linker DNA, and a few also appear to bind directly to the nucleosome.  TRY PROBLEM 32
TRY PROBLEM 32
HIGHER-
The next level of chromatin structure is a series of loops of the 30-
CONCEPTS
The nucleosome consists of a core particle of eight histone proteins and the DNA that wraps around them. A single H1 histone associates with each nucleosome. Nucleosomes are separated by linker DNA. Nucleosomes fold to form a 30-
CHANGES IN CHROMATIN STRUCTURE Although eukaryotic DNA must be tightly packed to fit into the cell nucleus, it must also periodically unwind to undergo transcription and replication. One process that alters chromatin structure is acetylation. Enzymes called acetyltransferases attach acetyl groups to lysine amino acids on the histone tails. This modification reduces the positive charges that normally exist on lysine and destabilizes the nucleosome structure, so the histones hold the DNA less tightly. Other chemical modifications of the histone proteins, such as methylation and phosphorylation, also alter chromatin structure, as do special chromatin-
A number of other changes can also affect chromatin structure, including the addition of methyl groups to DNA bases (DNA methylation), use of variant histone proteins in the nucleosome, and the binding of proteins to DNA and chromatin. Although these changes do not alter the DNA sequence, they often have major effects on the expression of genes, which will be considered in more detail in Chapter 12.
EPIGENETIC CHANGES ASSOCIATED WITH CHROMATIN MODIFICATIONS Some changes in chromatin structure are retained through cell division, so that they are passed on to future generations of cells and, occasionally, even to future generations of organisms. Alterations of chromatin structure that are passed on to descendant cells or individuals are frequently referred to as epigenetic changes, or simply as epigenetics (see Chapter 4). For example, the agouti locus helps determine coat color in mice: parents that have identical DNA sequences at this locus but have different degrees of methylation on their DNA may give rise to offspring that have different coat colors (Figure 8.20). Such epigenetic changes have been observed in a number of organisms and are responsible for a variety of phenotypic effects. Unlike mutations, epigenetic changes do not alter the DNA sequence, are capable of being reversed, and are often influenced by environmental factors. One type of epigenetic change is genomic imprinting, in which an allele is differentially expressed depending on whether it is inherited from the maternal or paternal parent (see Chapter 4). Genomic imprinting is caused by differences in DNA methylation of oocytes and sperm. The DNA of sperm and oocytes is differentially methylated at various imprinting control regions in the course of gamete formation, and the different methylation patterns of paternal and maternal alleles are then maintained and passed on to all resulting cells in the zygote.

CONCEPTS
Epigenetic changes are alterations of chromatin or DNA structure that do not include changes in the base sequence, but are stable and are passed on to descendant cells or organisms. Some epigenetic changes result from differences in DNA methylation.