
37.4 Histone Acetylation Results in Chromatin Remodeling
We have seen that nuclear receptors respond to signal molecules by recruiting coactivators. Now we can ask, how do coactivators modulate transcriptional activity? Recall that the template for RNA synthesis in eukaryotes is not simply naked DNA; rather, it is a complex of DNA and histones called chromatin. Some proteins that stimulate transcription act to loosen the histone complex from the DNA, exposing additional DNA regions to the transcription machinery. One means by which loosening takes place is with the enzymatic attachment of acetyl groups to histones.
Metabolism in Context: Acetyl CoA Plays a Key Role in the Regulation of Transcription
So far in our study of biochemistry, we have seen acetyl CoA only in the context of intermediary metabolism, for instance, as a fuel for the citric acid cycle or as a precursor for fatty acid synthesis and for steroid synthesis. However, acetyl CoA also plays a role in the regulation of gene expression. Recall that citrate is transported out of mitochondria and is cleaved into oxaloacetate and acetyl CoA in the cytoplasm by ATP-
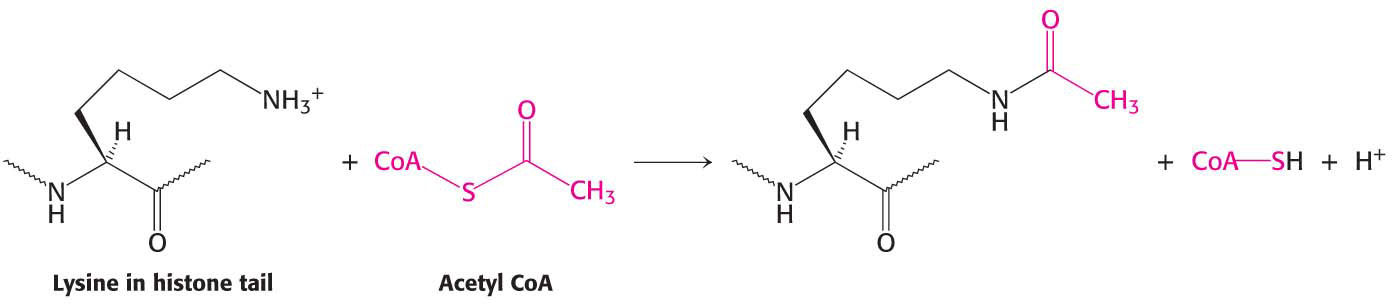
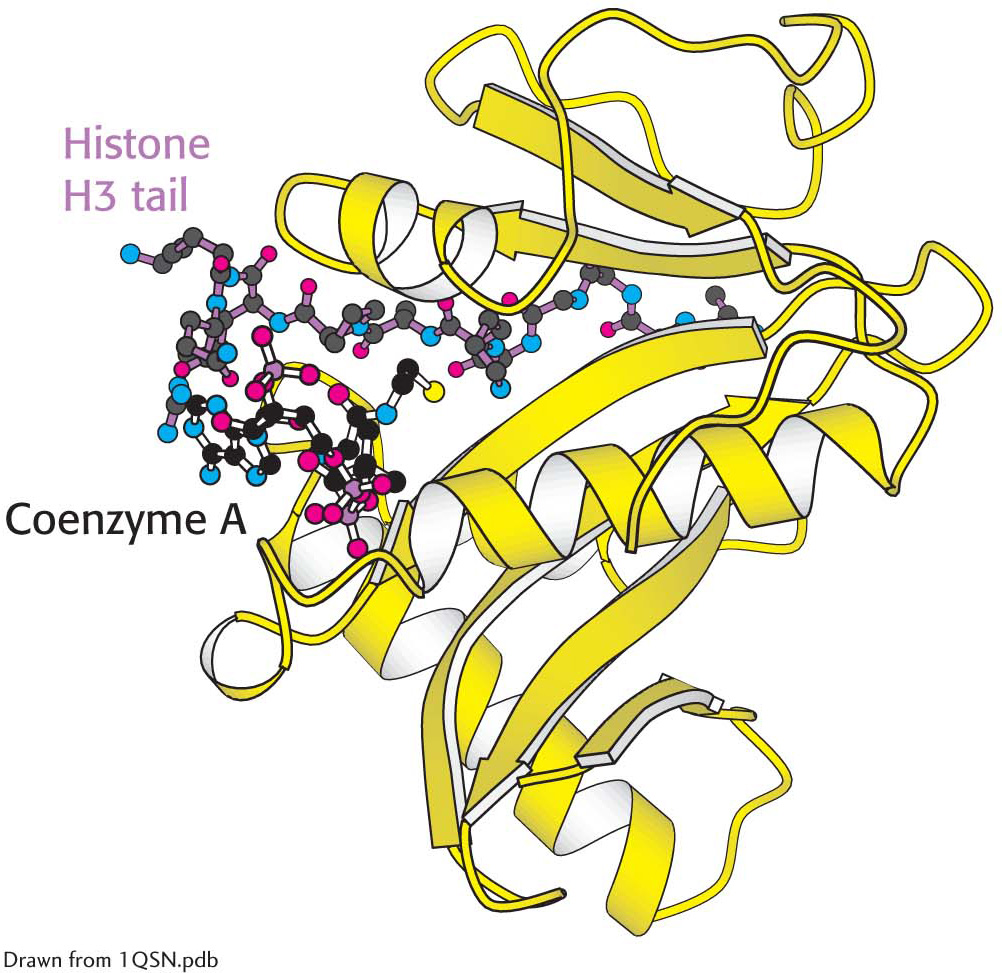
Enzymes that catalyze such reactions are called histone acetyltransferases (HATs). The histone tails are readily extended, so they can fit into the HAT active site and become acetylated (Figure 37.12).
What are the consequences of histone acetylation? Lysine bears a positively charged ammonium group at neutral pH. The addition of an acetyl group neutralizes the ammonium group to an amide group while adding a negative charge. This change dramatically reduces the affinity of the tail for DNA and decreases the affinity of the entire histone complex for DNA, loosening the histone complex from the DNA.
In addition, the acetylated lysine residues interact with a specific acetyllysine-
Bromodomains are also present in some components of large complexes known as chromatin-
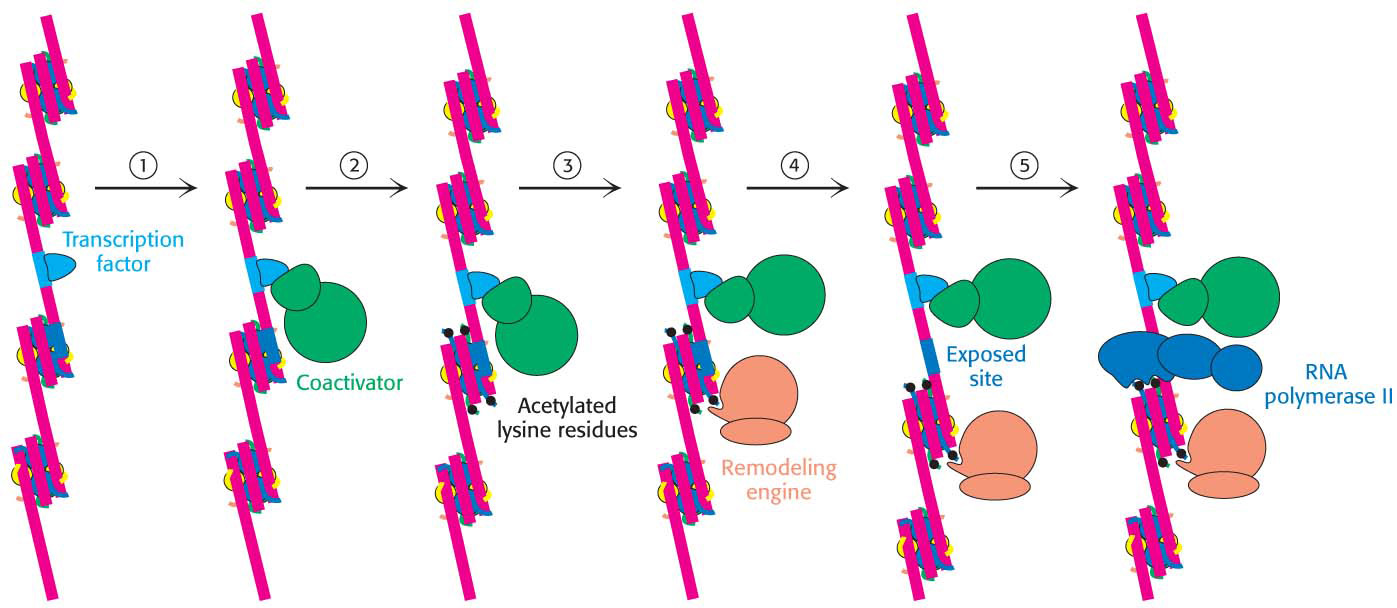
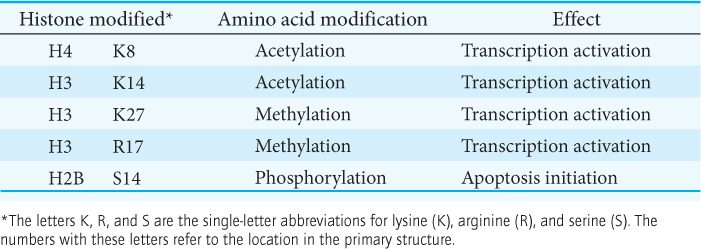
Recall that nuclear hormone receptors also include regions that interact with components of coactivators. Thus, two mechanisms of gene regulation can work in concert. The modification of histones and chromatin remodeling can open up regions of chromatin into which the transcription complex can be recruited through protein–
Histone Deacetylases Contribute to Transcriptional Repression
Just as in bacteria, some changes in a cell’s environment lead to the repression of genes that had been active. The modification of histone tails again plays an important role. However, in repression, a key reaction appears to be the deacetylation of acetylated lysine, catalyzed by specific histone deacetylase enzymes. Indeed, all covalent modifications of histone tails are reversible.
In many ways, the acetylation and deacetylation of lysine residues in histone tails is analogous to the phosphorylation and dephosphorylation of serine, threonine, and tyrosine residues in other stages of signaling processes. Like the addition of phosphoryl groups, the addition of acetyl groups can induce conformational changes and generate novel binding sites. Without a means of removing these groups, however, these signaling switches will become stuck in one position and lose their effectiveness. Like phosphatases, deacetylases help reset the switches.
