Chapter 30
Chapter 30
1. The Oxford English Dictionary defines translation as the action or process of turning from one language into another. Protein synthesis converts nucleic acid sequence information into amino acid sequence information.
2. An error frequency of 1 incorrect amino acid every 104 incorporations allows for the rapid and accurate synthesis of proteins as large as 1000 amino acids. Higher error rates would result in too many defective proteins. Lower error rates would likely slow the rate of protein synthesis without a significant gain in accuracy.
3. (i) Each is a single chain. (ii) They contain unusual bases. (iii) Approximately half of the bases are base-
4. First is the formation of the aminoacyl adenylate, which then reacts with the tRNA to form the aminoacyl-
5. Unique features are required so that the aminoacyl-
6. An activated amino acid is one linked to the appropriate tRNA.
7. (a) No; (b) no; (c) yes.
8. The ATP is cleaved to AMP and PPi. Consequently, a second ATP is required to convert AMP into ADP, the substrate for oxidative phosphorylation.
9. Amino acids larger than the correct amino acid cannot fit into the active site of the tRNA. Smaller but incorrect amino acids that become attached to the tRNA fit into the editing site and are cleaved from the tRNA.
10. Recognition sites on both faces of the tRNAs may be required to uniquely identify the 20 different tRNAs.
11. The first two bases in a codon form Watson–
12. Four bands: light, heavy, a hybrid of light 30S and heavy 50S, and a hybrid of heavy 30S and light 50S.
13. Two hundred molecules of ATP are converted into 200 AMP + 400 Pi to activate the 200 amino acids, which is equivalent to 400 molecules of ATP. One molecule of GTP is required for initiation, and 398 molecules of GTP are needed to form 199 peptide bonds.
14. The reading frame is a set of contiguous, nonoverlapping three-
15. A mutation caused by the insertion of an extra base can be suppressed by a tRNA that contains a fourth base in its anticodon. For example, UUUC rather than UUU is read as the codon for phenylalanine by a tRNA that contains 3′-AAAG-
16. One approach is to synthesize a tRNA that is acylated with a reactive amino acid analog. For example, bromoacetyl-
17. The sequence GAGGU is complementary to a sequence of five bases at the 3′ end of 16S rRNA and is located several bases upstream of an AUG start codon. Hence, this region is a start signal for protein synthesis. The replacement of G by A would be expected to weaken the interaction of this mRNA with the 16S rRNA and thereby diminish its effectiveness as an initiation signal. In fact, this mutation results in a 10-
18. The peptide would be Phe-
19. Proteins are synthesized from the amino to the carboxyl end on ribosomes, whereas they are synthesized in the reverse direction in the solid-
20. GTP is not hydrolyzed until aminoacyl-
21. The translation of an mRNA molecule can be blocked by antisense RNA, an RNA molecule with the complementary sequence. The antisense–
A44
22. (a) A5. (b) A5 > A4 > A3 > A2. (c) Synthesis is from the amino terminus to the carboxyl terminus.
23. These enzymes convert nucleic acid information into protein information by interpreting the tRNA and linking it to the proper amino acid.
24. The rate would fall because the elongation step requires that the GTP be hydrolyzed before any further elongation can take place.
25. Protein factors modulate the initiation of protein synthesis. The role of IF1 and IF3 is to prevent premature binding of the 30S and 50S ribosomal subunits, whereas IF2 delivers Met-
26. The signal sequence, signal-
27. The formation of peptide bonds, which in turn are powered by the hydrolysis of the aminoacyl-
28. The Shine–
29.
|
|
Bacteria |
Eukaryote |
|---|---|---|
|
Ribosome size |
70S |
80S |
|
mRNA |
Polycistronic |
Not polycistronic |
|
Initiation |
Shine– |
First AUG is used |
|
Protein factors |
Required |
Many more required |
|
Relation to transcription |
Translation can start before transcription is completed |
Transcription and translation are spatially separated |
|
First amino acid |
fMet |
Met |
30. The SRP binds to the signal sequence and inhibits further translation. The SRP ushers the inhibited ribosome to the ER, where it interacts with the SRP receptor (SR). The SRP–
31. The alternative would be to have a single ribosome translating a single mRNA molecule. The use of polysomes allows more protein synthesis per mRNA molecule in a given period of time and thus the production of more protein.
32. (a) 1, 2, 3, 5, 6, 10; (b) 1, 2, 7, 8; (c) 1, 4, 8, 9.
33. Transfer RNAs have roles in several recognition processes. A tRNA must be recognized by the appropriate aminoacyl-
34. The aminoacyl-
35. (a, d, and e) Type 2; (b, c, and f ) Type 1.
36. The error rates of DNA, RNA, and protein synthesis are of the order of 10−10, 10−5, and 10−4, respectively, per nucleotide (or amino acid) incorporated. The fidelity of all three processes depends on the precision of base-
37. EF-
38. The α subunits of G proteins are inhibited by a similar mechanism in cholera and whooping cough (Section 14.5).
39. Glu-
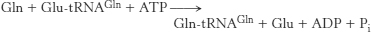
Glu-
40. The primary structure determines the three-
41. (a) eIF-
(b) To firmly establish that the effect of eIF-
(c) Half-
(d) eIF-
(e) The results in graph C suggest that eIF-
42. (a) The three peaks represent, from left to right, the 40S ribosomal subunit, the 60S ribosomal subunit, and the 80S ribosome.
(b) Not only are ribosomal subunits and the 80S ribosome present, but polysomes of various lengths also are apparent. The individual peaks in the polysome region represent polysomes of discrete length.
(c) The treatment significantly inhibited the number of polysomes while increasing the number of free ribosomal subunits. This outcome could be due to inhibited protein-