25.2Purine Bases Can Be Synthesized de Novo or Recycled by Salvage Pathways
Purine Bases Can Be Synthesized de Novo or Recycled by Salvage Pathways
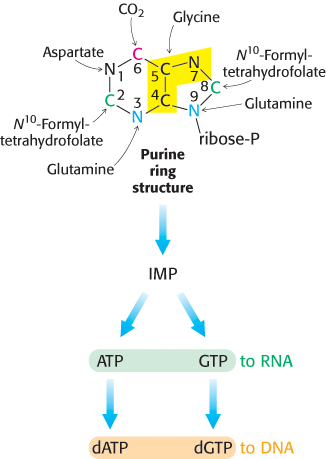
Like pyrimidine nucleotides, purine nucleotides can be synthesized de novo or by a salvage pathway. When synthesized de novo, purine synthesis begins with simple starting materials such as amino acids and bicarbonate (Figure 25.5). Unlike the bases of pyrimidines, the purine bases are assembled already attached to the ribose ring. Alternatively, purine bases, released by the hydrolytic degradation of nucleic acids and nucleotides, can be salvaged and recycled. Purine salvage pathways are especially noted for the energy that they save and the remarkable effects of their absence.
749
The purine ring system is assembled on ribose phosphate
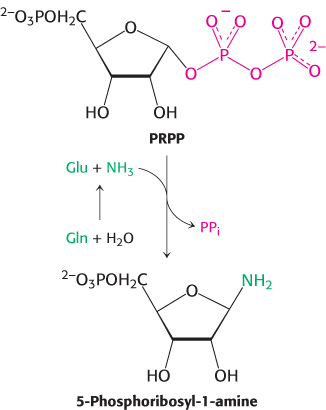
De novo purine biosynthesis, like pyrimidine biosynthesis, requires PRPP but, for purines, PRPP provides the foundation on which the bases are constructed step by step. The initial step is the displacement of pyrophosphate by ammonia, rather than by a preassembled base, to produce 5-
Glutamine phosphoribosyl amidotransferase catalyzes this reaction, which is the committed step in purine regulation. This enzyme comprises two domains: the first is homologous to the phosphoribosyltransferases in purine salvage pathways, whereas the second produces ammonia from glutamine by hydrolysis. However, this glutamine-
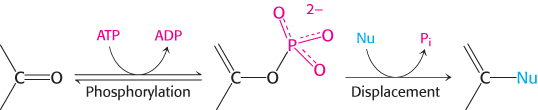
The purine ring is assembled by successive steps of activation by phosphorylation followed by displacement
 Nine additional steps are required to assemble the purine ring. Remarkably, the first six steps are analogous reactions. Most of these steps are catalyzed by enzymes with ATP-
Nine additional steps are required to assemble the purine ring. Remarkably, the first six steps are analogous reactions. Most of these steps are catalyzed by enzymes with ATP-
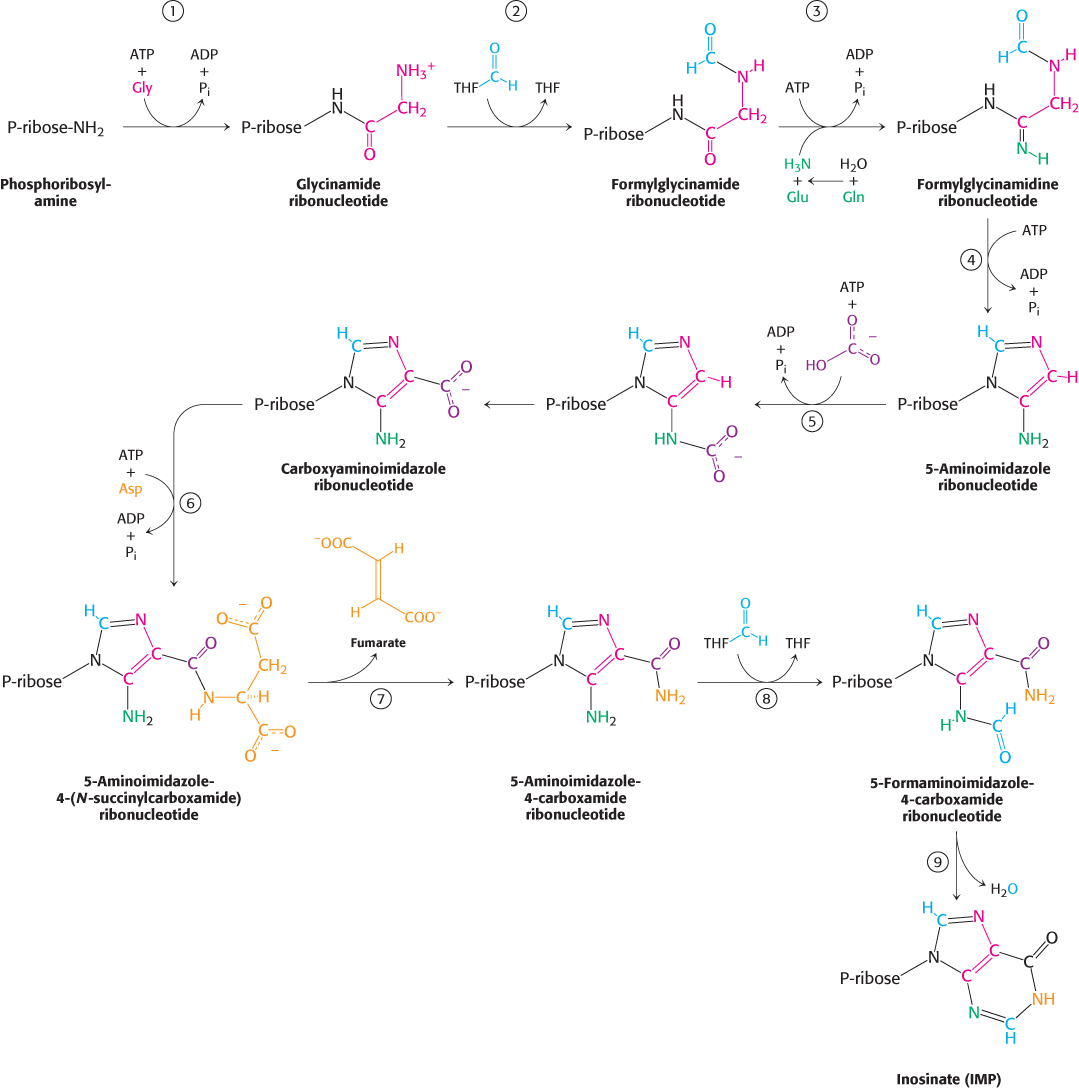
De novo purine biosynthesis proceeds as shown in Figure 25.6. Table 25.2 lists the enzymes that catalyze each step of the reaction.
|
Step |
Enzyme |
|---|---|
|
1 |
Glycinamide ribonucleotide (GAR) synthetase |
|
2 |
GAR transformylase |
|
3 |
Formylglycinamidine synthase |
|
4 |
Aminoimidazole ribonucleotide synthetase |
|
5 |
Carboxyaminoimidazole ribonucleotide synthetase |
|
6 |
Succinylaminoimidazole carboxamide ribonucleotide synthetase |
|
7 |
Adenylosuccinate lyase |
|
8 |
Aminoimidazole carboxamide ribonucleotide transformylase |
|
9 |
Inosine monophosphate cyclohydrolyase |
The carboxylate group of a glycine residue is activated by phosphorylation and then coupled to the amino group of phosphoribosylamine. A new amide bond is formed, and the amino group of glycine is free to act as a nucleophile in the next step.
N10-formyltetrahydrofolate donates a formyl moiety to this amino group to form formylglycinamide ribonucleotide.
750
The inner carbonyl group is activated by phosphorylation and then converted into an amidine by the addition of ammonia derived from glutamine.
The product of this reaction, formylglycinamidine ribonucleotide, cyclizes to form the five-
membered imidazole ring found in purines. Although this cyclization is likely to be favorable thermodynamically, a molecule of ATP is consumed to ensure irreversibility. The familiar pattern is repeated: a phosphoryl group from the ATP molecule activates the carbonyl group and is displaced by the nitrogen atom attached to the ribose molecule. Cyclization is thus an intramolecular reaction in which the nucleophile and phosphate- activated carbon atom are present within the same molecule. In higher eukaryotes, the enzymes catalyzing steps 1, 2, and 4 (Table 25.2) are components of a single polypeptide chain. 751
Bicarbonate is activated by phosphorylation and then attacked by the exocyclic amino group. The product of the reaction in step 5 rearranges to transfer the carboxylate group to the imidazole ring. Interestingly, mammals do not require ATP for this step; bicarbonate apparently attaches directly to the exocyclic amino group and is then transferred to the imidazole ring.
The imidazole carboxylate group is phosphorylated again and the phosphate group is displaced by the amino group of aspartate. Once again, in higher eukaryotes, the enzymes catalyzing steps 5 and 6 (Table 25.2) share a single polypeptide chain.
Fumarate, an intermediate in the citric acid cycle, is eliminated, leaving the nitrogen atom from aspartate joined to the imidazole ring. The use of aspartate as an amino-
group donor and the concomitant release of fumarate are reminiscent of the conversion of citrulline into arginine in the urea cycle, and these steps are catalyzed by homologous enzymes in the two pathways (Section 23.4). A formyl group from N10-formyltetrahydrofolate is added to this nitrogen atom to form a final 5-
formaminoimidazole- 4- carboxamide ribonucleotide. 5-
Formaminoimidazole- 4- carboxamide ribonucleotide cyclizes with the loss of water to form inosinate.
Many of the intermediates in the de novo purine biosynthesis pathway degrade rapidly in water. Their instability in water suggests that the product of one enzyme must be channeled directly to the next enzyme along the pathway. Recent evidence shows that the enzymes do indeed form complexes when purine synthesis is required.
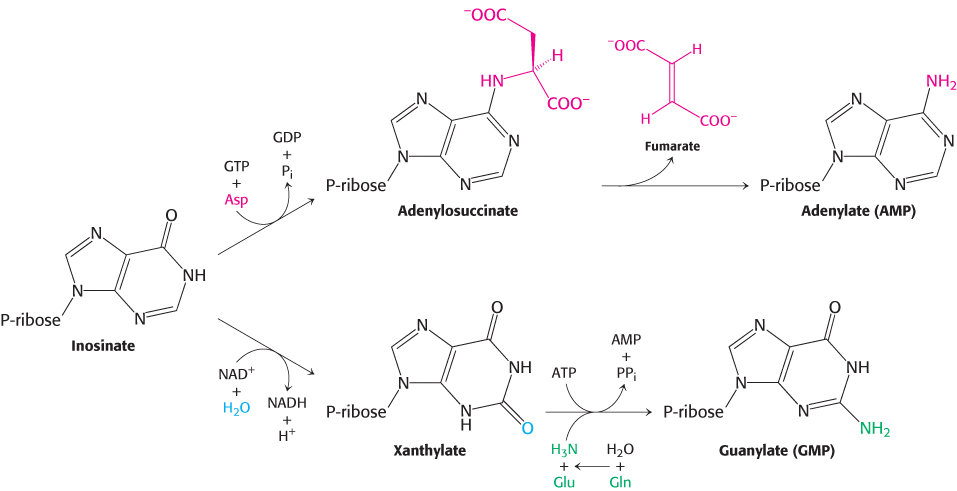
AMP and GMP are formed from IMP
A few steps convert inosinate into either AMP or GMP (Figure 25.7). Adenylate is synthesized from inosinate by the substitution of an amino group for the carbonyl oxygen atom at C-
752
Guanylate is synthesized by the oxidation of inosinate to xanthylate (XMP), followed by the incorporation of an amino group at C-
Enzymes of the purine synthesis pathway associate with one another in vivo
Biochemists believe that the enzymes of many metabolic pathways, such as glycolysis and the citric acid cycle, are physically associated with one another. Such associations would increase the efficiency of pathways by facilitating the movement of the product of one enzyme to the active site of the next enzyme in the pathway. The evidence for such associations comes primarily from experiments in which one component of a pathway, carefully isolated from the cell, is found to be bound to other components of the pathway. However, these observations raise the question, do enzymes associate with one another in vivo or do they spuriously associate during the isolation procedure? Recent in vivo evidence shows that the enzymes of the purine synthesis pathway associate with one another when purine synthesis is required. Various enzymes of the pathway were fused with the green fluorescent protein (Figure 2.65) and transfected into cells. When cells were grown in the presence of purine, the GFP was spread diffusely throughout the cytoplasm (Figure 25.8A). When the cells were switched to growth media without purines, purine synthesis began and the enzymes became associated with one another, forming complexes dubbed purinosomes (Figure 25.8B). The experiments were repeated with other enzymes of the purine synthesis pathway bearing the GFP, and the results were the same: purine synthesis occurs when the enzymes form the purinosomes. What actually causes complex formation? While the results are not yet established, it appears that several G-

Salvage pathways economize intracellular energy expenditure
As we have seen, the de novo synthesis of purines requires a substantial investment of ATP. Purine salvage pathways provide a more economical means of generating purines. Free purine bases, derived from the turnover of nucleotides or from the diet, can be attached to PRPP to form purine nucleoside monophosphates, in a reaction analogous to the formation of orotidylate. Two salvage enzymes with different specificities recover purine bases. Adenine phosphoribosyltransferase catalyzes the formation of adenylate (AMP):

whereas hypoxanthine-
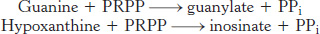
753