25.4Key Steps in Nucleotide Biosynthesis Are Regulated by Feedback Inhibition
Key Steps in Nucleotide Biosynthesis Are Regulated by Feedback Inhibition
Nucleotide biosynthesis is regulated by feedback inhibition in a manner similar to the regulation of amino acid biosynthesis (Section 24.3). These regulatory pathways ensure that the various nucleotides are produced in the required quantities.
Pyrimidine biosynthesis is regulated by aspartate transcarbamoylase
Aspartate transcarbamoylase, one of the key enzymes for the regulation of pyrimidine biosynthesis in bacteria, was described in detail in Chapter 10. Recall that ATCase is inhibited by CTP, the final product of pyrimidine biosynthesis, and stimulated by ATP.

Carbamoyl phosphate synthetase II is also a site of feedback inhibition in both prokaryotes and eukaryotes.
The synthesis of purine nucleotides is controlled by feedback inhibition at several sites
The regulatory scheme for purine nucleotides is more complex than that for pyrimidine nucleotides (Figure 25.15).
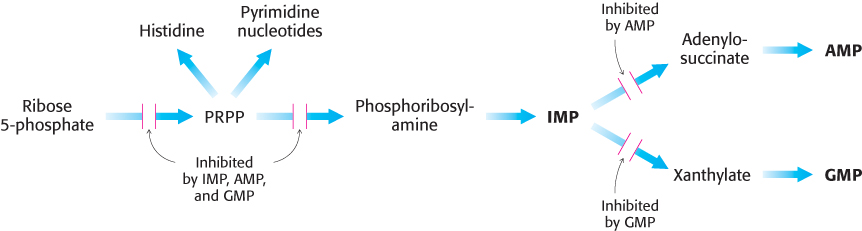
The committed step in purine nucleotide biosynthesis is the conversion of PRPP into phosphoribosylamine by glutamine phosphoribosyl amidotransferase. This important enzyme is feedback-
inhibited by many purine ribonucleotides. It is noteworthy that AMP and GMP, the final products of the pathway, synergistically inhibit the amidotransferase. 759
Inosinate is the branch point in the synthesis of AMP and GMP. The reactions leading away from inosinate are sites of feedback inhibition. AMP inhibits the conversion of inosinate into adenylosuccinate, its immediate precursor. Similarly, GMP inhibits the conversion of inosinate into xanthylate, its immediate precursor.
As already noted, GTP is a substrate in the synthesis of AMP, whereas ATP is a substrate in the synthesis of GMP (Figure 25.7). This reciprocal substrate relation tends to balance the synthesis of adenine and guanine ribonucleotides.
Note that the synthesis of PRPP by PRPP synthetase is highly regulated even though it is not the committed step in purine synthesis. Mutations have been identified in PRPP synthetase that result in a loss of allosteric response to nucleotides without any effect on catalytic activity of the enzyme. A consequence of this mutation is an overabundance of purine nucleotides that can result in gout, a pathological condition discussed later in the chapter.
The synthesis of deoxyribonucleotides is controlled by the regulation of ribonucleotide reductase
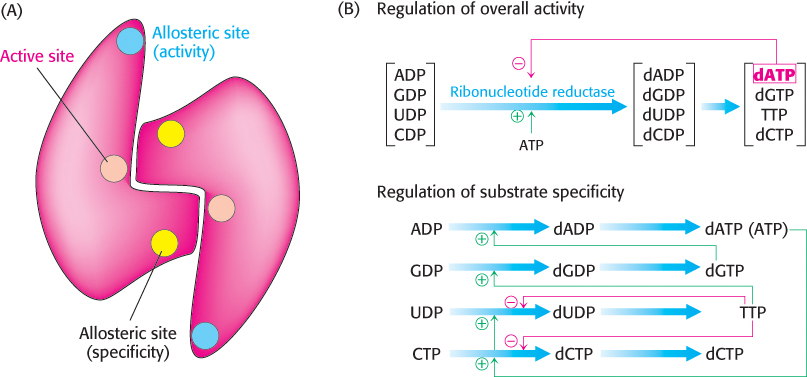
The reduction of ribonucleotides to deoxyribonucleotides is precisely controlled by allosteric interactions. Each polypeptide of the R1 subunit of the aerobic E. coli ribonucleotide reductase contains two allosteric sites: one of them controls the overall activity of the enzyme, and the other regulates substrate specificity (Figure 25.16A). The overall catalytic activity of ribonucleotide reductase is diminished by the binding of dATP, which signals an abundance of deoxyribonucleotides (Figure 25.16B). The binding of ATP reverses this feedback inhibition. The binding of dATP or ATP to the substrate-
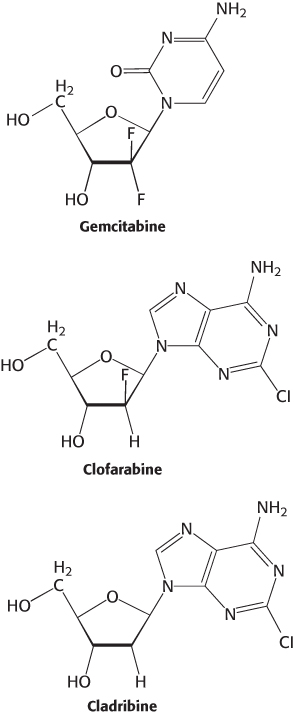
 Ribonucleotide reductase is an attractive target for cancer therapy and a number of clinically approved anticancer drugs mimic substrates and regulators of the enzyme. The pyrimidine analog gemcitabine, once converted to the diphosphate form in vivo, becomes a suicide inhibitor of the reductase used to treat advanced pancreatic cancer. The purine analogs clofarabine and cladribine, upon conversion to their triphosphate forms in vivo, are dATP analogs and act as allosteric inhibitors of the enzyme. Clofarabine is used to treat pediatric acute myeloid leukemia while cladribine is effective against some forms of chronic lymphoid leukemia.
Ribonucleotide reductase is an attractive target for cancer therapy and a number of clinically approved anticancer drugs mimic substrates and regulators of the enzyme. The pyrimidine analog gemcitabine, once converted to the diphosphate form in vivo, becomes a suicide inhibitor of the reductase used to treat advanced pancreatic cancer. The purine analogs clofarabine and cladribine, upon conversion to their triphosphate forms in vivo, are dATP analogs and act as allosteric inhibitors of the enzyme. Clofarabine is used to treat pediatric acute myeloid leukemia while cladribine is effective against some forms of chronic lymphoid leukemia.
760