Regulatory Strategies
CHAPTER
10
285
OUTLINE
The activity of enzymes must often be regulated so that they function at the proper time and place. This regulation is essential for coordination of the vast array of biochemical processes taking place at any instant in an organism. Enzymatic activity is regulated in five principal ways:
286
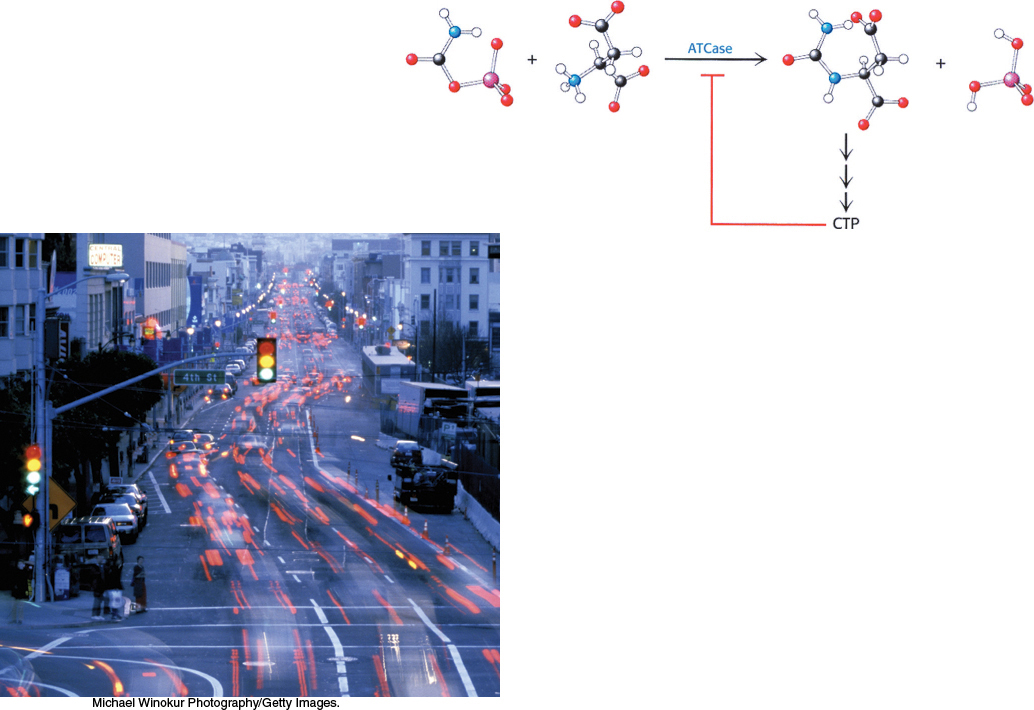
Allosteric Control. Allosteric proteins contain distinct regulatory sites and multiple functional sites. The binding of small signal molecules at regulatory sites controls the activity of these proteins. Moreover, allosteric proteins show the property of cooperativity: activity at one functional site affects the activity at others. Proteins displaying allosteric control are thus information transducers: their activity can be modified in response to signal molecules or to information shared among active sites. This chapter examines one of the best-
understood allosteric proteins: the enzyme aspartate transcarbamoylase (ATCase). Catalysis by aspartate transcarbamoylase of the first step in pyrimidine biosynthesis is inhibited by cytidine triphosphate, the final product of that biosynthesis, in an example of feedback inhibition. We have already examined an allosteric protein— hemoglobin, the oxygen transport protein in the blood (Chapter 7). Multiple Forms of Enzymes. Isozymes, or isoenzymes, provide an avenue for varying regulation of the same reaction to meet the specific physiological needs in the particular tissue at a particular time. Isozymes are homologous enzymes within a single organism that catalyze the same reaction but differ slightly in structure and more obviously in KM and Vmax values as well as in regulatory properties. Often, isozymes are expressed in a distinct tissue or organelle or at a distinct stage of development.
Reversible Covalent Modification. The catalytic properties of many enzymes are markedly altered by the covalent attachment of a modifying group, commonly a phosphoryl group. ATP serves as the phosphoryl donor in these reactions, which are catalyzed by protein kinases. The removal of phosphoryl groups by hydrolysis is catalyzed by protein phosphatases. This chapter considers the structure, specificity, and control of protein kinase A (PKA), a ubiquitous eukaryotic enzyme that regulates diverse target proteins.
Proteolytic Activation. The enzymes controlled by some of these regulatory mechanisms cycle between active and inactive states. A different regulatory strategy is used to irreversibly convert an inactive enzyme into an active one. Many enzymes are activated by the hydrolysis of a few peptide bonds or even one such bond in inactive precursors called zymogens or proenzymes. This regulatory mechanism generates digestive enzymes such as chymotrypsin, trypsin, and pepsin. Blood clotting is due to a remarkable cascade of zymogen activations. Active digestive and clotting enzymes are switched off by the irreversible binding of specific inhibitory proteins that are irresistible lures to their molecular prey.
Controlling the Amount of Enzyme Present. Enzyme activity can also be regulated by adjusting the amount of enzyme present. This important form of regulation usually takes place at the level of transcription. We will consider the control of gene transcription in Chapters 29, 30, and 31.
To begin, we will consider the principles of allostery by examining the enzyme aspartate transcarbamoylase.