28.1DNA Replication Proceeds by the Polymerization of Deoxyribonucleoside Triphosphates Along a Template
DNA Replication Proceeds by the Polymerization of Deoxyribonucleoside Triphosphates Along a Template
The base sequences of newly synthesized DNA must faithfully match the sequences of parent DNA. To achieve faithful replication, each strand within the parent double helix acts as a template for the synthesis of a new DNA strand with a complementary sequence. The building blocks for the synthesis of the new strands are deoxyribonucleoside triphosphates. They are added, one at a time, to the 3′ end of an existing strand of DNA.
Although this reaction is in principle quite simple, it is significantly complicated by specific features of the DNA double helix. First, the two strands of the double helix run in opposite directions. Because DNA strand synthesis always proceeds in the 5′-to-
829
DNA polymerases require a template and a primer
Primer
The initial segment of a polymer that is to be extended on which elongation depends.
Template
A sequence of DNA or RNA that directs the synthesis of a complementary sequence.
DNA polymerases catalyze the formation of polynucleotide chains. Each incoming nucleoside triphosphate first forms an appropriate base pair with a base in the template. Only then does the DNA polymerase link the incoming base with the predecessor in the chain. Thus, DNA polymerases are template-
DNA polymerases add nucleotides to the 3′ end of a polynucleotide chain. The polymerase catalyzes the nucleophilic attack by the 3′-hydroxyl-
All DNA polymerases have structural features in common
The three-
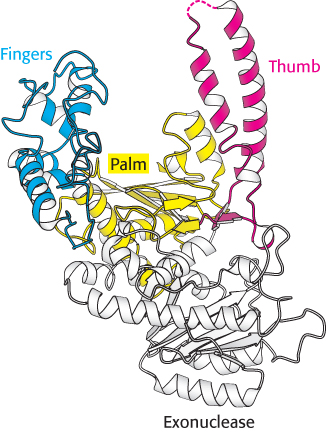
 FIGURE 28.4 DNA polymerase structure. The first DNA polymerase structure determined was that of a fragment of E. coli DNA polymerase I called the Klenow fragment. Notice that, like other DNA polymerases, the polymerase unit resembles a right hand with fingers (blue), palm (yellow), and thumb (red). The Klenow fragment also includes an exonuclease domain that removes incorrect nucleotide bases.
FIGURE 28.4 DNA polymerase structure. The first DNA polymerase structure determined was that of a fragment of E. coli DNA polymerase I called the Klenow fragment. Notice that, like other DNA polymerases, the polymerase unit resembles a right hand with fingers (blue), palm (yellow), and thumb (red). The Klenow fragment also includes an exonuclease domain that removes incorrect nucleotide bases.
 DNA polymerases are remarkably similar in overall shape, although they differ substantially in detail. At least five structural classes have been identified; some of them are clearly homologous, whereas others appear to be the products of convergent evolution. In all cases, the finger and thumb domains wrap around DNA and hold it across the enzyme’s active site, which comprises residues primarily from the palm domain. Furthermore, all DNA polymerases use similar strategies to catalyze the polymerase reaction, making use of a mechanism in which two metal ions take part.
DNA polymerases are remarkably similar in overall shape, although they differ substantially in detail. At least five structural classes have been identified; some of them are clearly homologous, whereas others appear to be the products of convergent evolution. In all cases, the finger and thumb domains wrap around DNA and hold it across the enzyme’s active site, which comprises residues primarily from the palm domain. Furthermore, all DNA polymerases use similar strategies to catalyze the polymerase reaction, making use of a mechanism in which two metal ions take part.
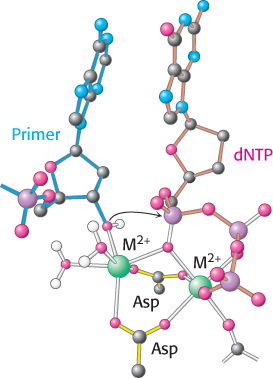
Two bound metal ions participate in the polymerase reaction
Like all enzymes with nucleoside triphosphate substrates, DNA polymerases require metal ions for activity. Examination of the structures of DNA polymerases with bound substrates and substrate analogs reveals the presence of two metal ions in the active site. One metal ion binds both the deoxynucleoside triphosphate (dNTP) and the 3′-hydroxyl group of the primer, whereas the other interacts only with the dNTP (Figure 28.5). The two metal ions are bridged by the carboxylate groups of two aspartate residues in the palm domain of the polymerase. These side chains hold the metal ions in the proper positions and orientations. The metal ion bound to the primer activates the 3′-hydroxyl group of the primer, facilitating its attack on the α phosphoryl group of the dNTP substrate in the active site. The two metal ions together help stabilize the negative charge that accumulates on the pentacoordinate transition state. The metal ion initially bound to dNTP stabilizes the negative charge on the pyrophosphate product.
 P bond.
P bond.
830
The specificity of replication is dictated by complementarity of shape between bases
DNA must be replicated with high fidelity. Each base added to the growing chain should, with high probability, be the Watson–
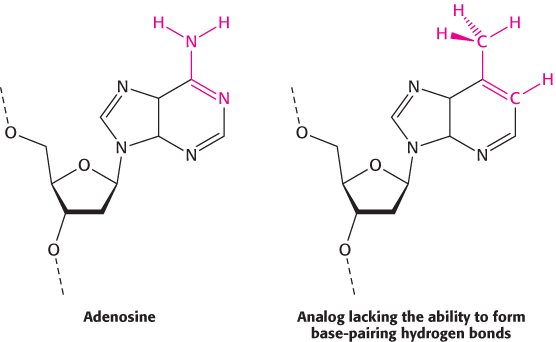
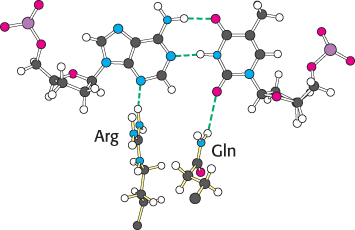
An examination of the crystal structures of various DNA polymerases reveals why shape complementarity is so important. First, residues of the enzyme form hydrogen bonds with the minor-
Second, DNA polymerases close down around the incoming dNTP (Figure 28.8). The binding of a deoxyribonucleoside triphosphate into the active site of a DNA polymerase triggers a conformational change: the finger domain rotates to form a tight pocket into which only a properly shaped base pair will readily fit. Many of the residues lining this pocket are important to ensure the efficiency and fidelity of DNA synthesis. For example, mutation of a conserved tyrosine residue that forms part of the pocket results in a polymerase that is approximately 40 times as error prone as the parent polymerase.
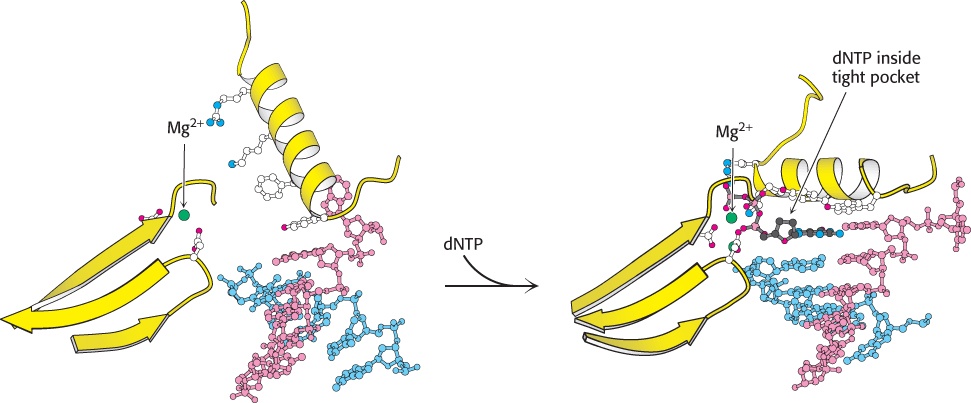
 FIGURE 28.8 Shape selectivity. The binding of a deoxyribonucleoside triphosphate (dNTP) to DNA polymerase induces a conformational change, generating a tight pocket for the base pair consisting of the dNTP and its partner on the template strand. Such a conformational change is possible only when the dNTP corresponds to the Watson–
FIGURE 28.8 Shape selectivity. The binding of a deoxyribonucleoside triphosphate (dNTP) to DNA polymerase induces a conformational change, generating a tight pocket for the base pair consisting of the dNTP and its partner on the template strand. Such a conformational change is possible only when the dNTP corresponds to the Watson–831
An RNA primer synthesized by primase enables DNA synthesis to begin

DNA polymerases cannot initiate DNA synthesis without a primer, a section of nucleic acid having a free 3′ end that forms a double helix with the template. How is this primer formed? An important clue came from the observation that RNA synthesis is essential for the initiation of DNA synthesis. In fact, RNA primes the synthesis of DNA. An RNA polymerase called primase synthesizes a short stretch of RNA (about five nucleotides) that is complementary to one of the template DNA strands (Figure 28.9). Primase, like other RNA polymerases, can initiate synthesis without a primer. After DNA synthesis has been initiated, the short stretch of RNA is removed by hydrolysis and replaced by DNA.
One strand of DNA is made continuously, whereas the other strand is synthesized in fragments
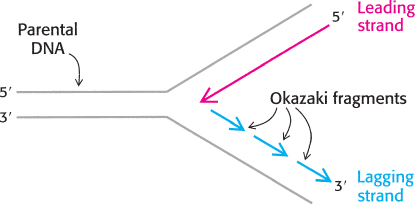
Both strands of parental DNA serve as templates for the synthesis of new DNA. The site of DNA synthesis is called the replication fork because the complex formed by the newly synthesized daughter helices arising from the parental duplex resembles a two-
This dilemma was resolved by Reiji Okazaki, who found that a significant proportion of newly synthesized DNA exists as small fragments. These units of about a thousand nucleotides (called Okazaki fragments) are present briefly in the vicinity of the replication fork (Figure 28.10).
832
As replication proceeds, these fragments become covalently joined through the action of the enzyme DNA ligase to form a continuous daughter strand. The other new strand is synthesized continuously. The strand formed from Okazaki fragments is termed the lagging strand, whereas the one synthesized without interruption is the leading strand. The discontinuous assembly of the lagging strand enables 5′ → 3′ polymerization at the nucleotide level to give rise to overall growth in the 3′ → 5′ direction.
DNA ligase joins ends of DNA in duplex regions
The joining of Okazaki fragments requires an enzyme that catalyzes the joining of the ends of two DNA chains. The existence of circular DNA molecules also points to the existence of such an enzyme. In 1967, scientists in several laboratories simultaneously discovered DNA ligase. This enzyme catalyzes the formation of a phosphodiester linkage between the 3′-hydroxyl group at the end of one DNA chain and the 5′-phosphoryl group at the end of the other (Figure 28.11). An energy source is required to drive this thermodynamically uphill reaction. In eukaryotes and archaea, ATP is the energy source. In bacteria, NAD+ typically plays this role.
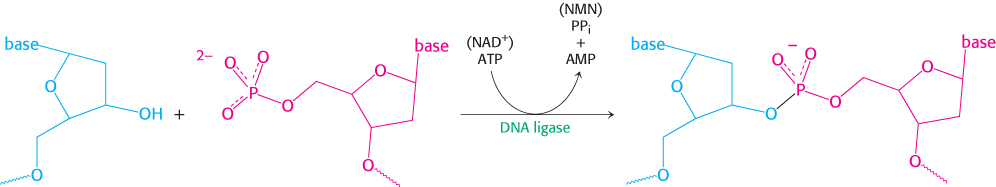
DNA ligase cannot link two molecules of single-
The separation of DNA strands requires specific helicases and ATP hydrolysis
For a double-
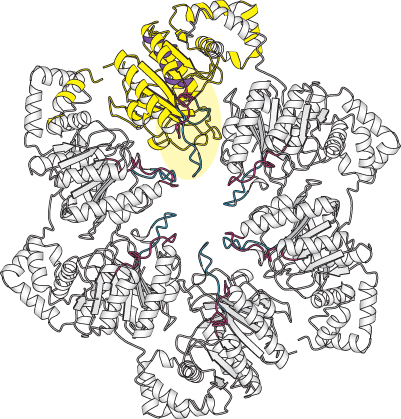
 FIGURE 28.12 Helicase structure. The structure of the hexameric helicase from bacteriophage T7. One of the six subunits is shown in yellow with the P-
FIGURE 28.12 Helicase structure. The structure of the hexameric helicase from bacteriophage T7. One of the six subunits is shown in yellow with the P-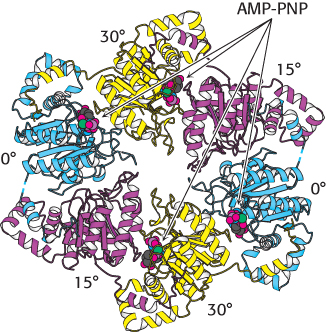
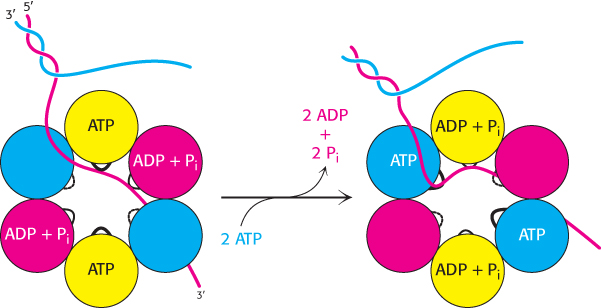
 FIGURE 28.13 Helicase asymmetry. The structure of the T7 helicase complexes with the ATP analog AMP-
FIGURE 28.13 Helicase asymmetry. The structure of the T7 helicase complexes with the ATP analog AMP-Helicases are a large and diverse family of enzymes taking part in many biological processes. The helicases in DNA replication are typically oligomers containing six subunits that form a ring structure. The structure of one such helicase, that from bacteriophage T7, has been a source of considerable insight into the helicase mechanism (Figure 28.12). Each of the subunits within this hexameric structure has a core structure that includes a P-
833
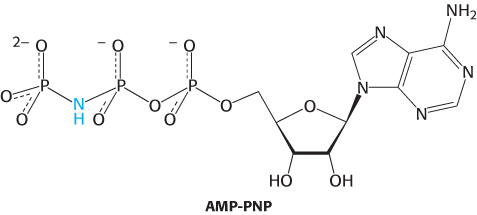
The AMP-
These observations are consistent with the following mechanism for the helicase (Figure 28.14). Only a single strand of DNA can fit through the center on the ring. This single strand binds to loops on two adjacent subunits, one of which has bound ATP and the other of which has bound ADP + Pi. The binding of ATP to the domains that initially had no bound nucleotides leads to a conformational change within the entire hexamer, leading to the release of ADP + Pi from two subunits and the binding of the single-