30.6
Ribosomes Bound to the Endoplasmic Reticulum Manufacture Secretory and Membrane Proteins
Not all newly synthesized proteins are destined to function in the cytoplasm. Eukaryotic cells can direct proteins to internal sites such as mitochondria, the nucleus and the endoplasmic reticulum, a process called protein targeting or protein sorting. How is sorting accomplished? There are two general mechanisms by which sorting takes place. In one mechanism, the protein is synthesized in the cytoplasm, and then the completed protein is delivered to its intracellular location posttranslationally. Proteins destined for the nucleus, chloroplasts, mitochondria, and peroxisomes are delivered by this general process. The other mechanism, termed the secretory pathway, directs proteins into the endoplasmic reticulum (ER), the extensive membrane system that comprises about half the total membrane of a cell, cotranslationally—that is, while the protein is being synthesized. Approximately 30% of all proteins are sorted by the secretory pathway, including secreted proteins, residents of the ER, the Golgi complex, lysosomes, and integral membrane proteins of these organelles as well as integral plasma-membrane proteins. We will focus our attention on the secretory pathway only.
In eukaryotic cells, a ribosome remains free in the cytoplasm unless it is directed to the ER. The region that binds ribosomes is called the rough ER because of its studded appearance in contrast with the smooth ER, which is devoid of ribosomes (Figure 30.33).
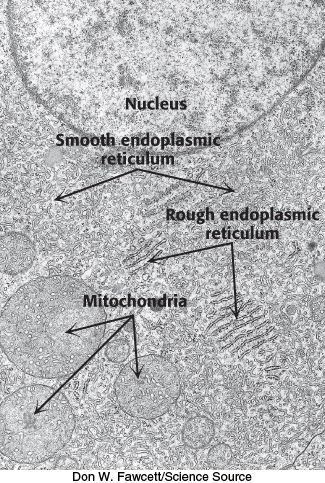
FIGURE 30.33Ribosomes are bound to the endoplasmic reticulum. In this electron micrograph, ribosomes appear as small black dots binding to the cytoplasmic side of the endoplasmic reticulum to give a rough appearance. In contrast, the smooth endoplasmic reticulum is devoid of ribosomes.
Signal sequences mark proteins for translocation across the endoplasmic reticulum membrane
The synthesis of proteins destined to leave the cell or become embedded in the plasma membrane begins on a free ribosome but, shortly after synthesis begins, it is halted until the ribosome is directed to the cytoplasmic side of the ER. When the ribosome docks with the ER membrane, protein synthesis begins again. As the newly forming peptide chain exits the ribosome, it is transported, cotranslationally, through the membrane into the lumen of the ER. The translocation consists of four components.
1. The Signal Sequence. The signal sequence is a sequence of 9 to 12 hydrophobic amino acid residues, sometimes containing positively charged amino acids (Figure 30.34). This sequence, which adopts an α-helical structure, is usually near the amino terminus of the nascent polypeptide chain. The presence of the signal sequence identifies the nascent peptide as one that must cross the ER membrane. Some signal sequences are maintained in the mature protein, whereas others are cleaved by a signal peptidase on the lumenal side of the ER membrane.

FIGURE 30.34Amino-terminal signal sequences of some eukaryotic secretory and plasma-membrane proteins. The hydrophobic core (yellow) is preceded by basic residues (blue) and followed by a cleavage site (red) for signal peptidase.
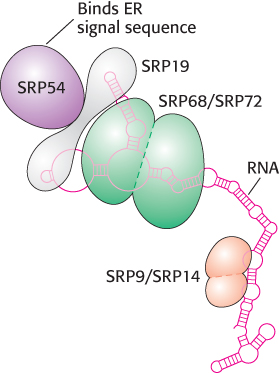
FIGURE 30.35The signal-recognition particle. The signal-recognition particle (SRP) consists of six proteins and one 300-nucleotide RNA molecule. The RNA has a complex structure with many double-helical stretches punctuated by single-stranded regions, shown as circles.
[Information from H. Lodish et al., Molecular Cell Biology, 5th ed. (W. H. Freeman and Company, 2004). Data from K. Strub et al., Mol. Cell Biol. 11:3949–3959, 1991, and S. High and B. Dobberstein, J. Cell Biol. 113:229–233, 1991.]
2. The Signal-Recognition Particle (SRP). The signal-recognition particle recognizes the signal sequence and binds the sequence and the ribosome as soon as the signal sequence exits the ribosome. SRP then shepherds the ribosome and its nascent polypeptide chain to the ER membrane. SRP is a ribonucleoprotein consisting of a 7S RNA and six different proteins (Figure 30.35). One protein, SRP54, is a GTPase that is crucial for SRP function. SRP samples all ribosomes until it locates one exhibiting a signal sequence. After SRP is bound to the signal sequence, interactions between the ribosome and the SRP occlude the elongation-factor-binding site, thereby halting protein synthesis.
3. The SRP Receptor (SR). The SRP–ribosome complex diffuses to the ER, where SRP binds the SRP receptor, an integral membrane protein consisting of two subunits, SRα and SRβ. SRα is, like SRP54, a GTPase.
4. The Translocon. The SRP–SR complex delivers the ribosome to the translocation machinery, called the translocon, a multisubunit assembly of integral and peripheral membrane proteins. The translocon is a protein-conducting channel. This channel opens when the translocon and ribosome bind to each other. Protein synthesis resumes with the growing polypeptide chain passing through the translocon channel into the lumen of the ER.
The interactions of the components of the translocation machinery are shown in Figure 30.36. Both the SRP54 and the SRα subunits of SR must bind GTP to facilitate the formation of the SRP–SR complex. For the SRP–SR complex to then deliver the ribosome to the translocon, the two GTP molecules—one in SRP and the other in SR—are aligned in what is essentially an active site shared by the two proteins. The formation of the alignment is catalyzed by the 7S RNA of the SRP. After the ribosome has been passed along to the translocon, the GTPs are hydrolyzed, SRP and SR dissociate, and SRP is free to search for another signal sequence to begin the cycle anew. Thus, SRP acts catalytically. The signal peptidase, which is associated with the translocon in the lumen of the ER, removes the signal sequence from most proteins.
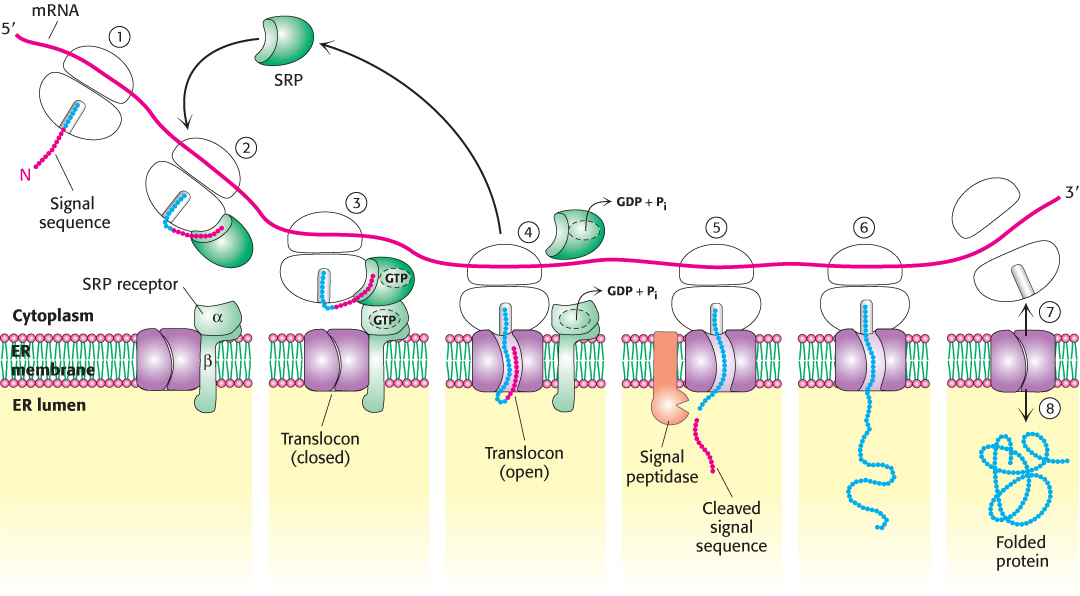
FIGURE 30.36The SRP targeting cycle. (1) Protein synthesis begins on free ribosomes. (2) After the signal sequence has exited the ribosome, it is bound by the SRP, and protein synthesis halts. (3) The SRP–ribosome complex docks with the SRP receptor in the ER membrane. (4) The SRP and the SRP receptor simultaneously hydrolyze bound GTPs. Protein synthesis resumes and the SRP is free to bind another signal sequence. (5) The signal peptidase may remove the signal sequence as it enters the lumen of the ER. (6) Protein synthesis continues as the protein is synthesized directly into the ER. (7) On completion of protein synthesis, the ribosome is released. (8) The protein tunnel in the translocon closes.
[Information from H. Lodish et al., Molecular Cell Biology, 5th ed. (W. H. Freeman and Company, 2004), Fig. 16.6.]
Transport vesicles carry cargo proteins to their final destination
As the proteins are synthesized, they fold to form their three-dimensional structures in the lumen of the ER. Some proteins are modified by the attachment of N-linked carbohydrates (Section 11.3). Finally, the proteins must be sorted and transported to their final destinations. Regardless of the destination, the principles of transport are the same. Transport is mediated by transport vesicles that bud off the ER (Figure 30.37). Transport vesicles from the ER carry their cargo (the proteins) to the Golgi complex, where the vesicles fuse and deposit the cargo inside the complex. There the cargo proteins are further modified—for instance, by the attachment of O-linked carbohydrates. From the Golgi complex, transport vesicles carry the cargo proteins to their final destinations, as shown in Figure 30.37.
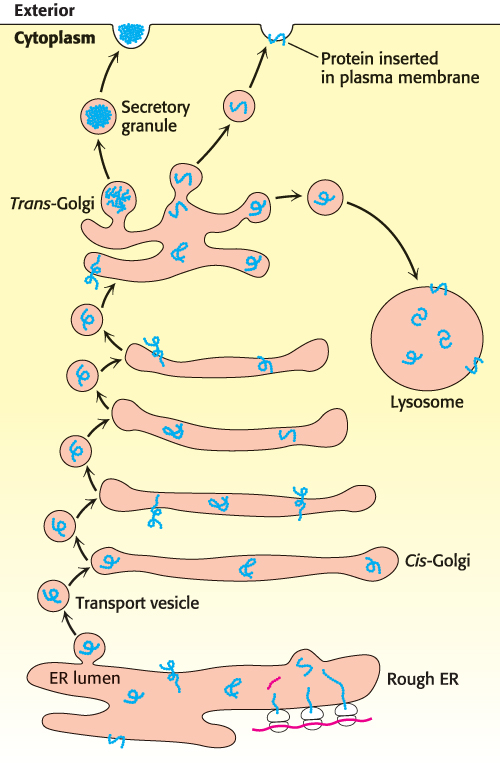
FIGURE 30.37Protein-sorting pathways. Newly synthesized proteins in the lumen of the ER are collected into membrane buds. These buds pinch off to form transport vesicles. The transport vesicles carry the cargo proteins to the Golgi complex, where the cargo proteins are modified. Transport vesicles then carry the cargo to the final destination as directed by the v-SNARE and t-SNARE proteins.
How does a protein end up at the correct destination? A newly synthesized protein will float inside the ER lumen until it binds to an integral membrane protein called a cargo receptor. This binding sequesters the cargo protein into a small region of the membrane that can subsequently form a membrane bud. The bud will carry the protein to a specific destination—plasma membrane, lysosome, or cell exterior. The key to ensuring that the protein reaches the proper destination is that the protein must bind to a receptor in the ER region associated with the protein’s destination. To ensure the proper match of protein with ER region, cargo receptors recognize various characteristics of the cargo protein, such as a particular amino acid sequence or an added carbohydrate.
The formation of buds is facilitated by the binding of coat proteins (COPs) to the cytoplasmic side of the bud. The coat proteins associate with one another to pinch off the vesicle. After the transport vesicle has formed and is released, the coat proteins are shed to reveal another integral protein called v-SNARE (“v” for vesicle). v-SNARE will bind to a particular t-SNARE (“t” for target) in the target membrane. This binding leads to the fusion of the transport vesicle to the target membrane, and the cargo is delivered. Thus, the assignment of identical v-SNARE proteins to the same region of the ER membrane causes an ER region to be associated with a particular destination.




