33.3
Photoreceptor Molecules in the Eye Detect Visible Light

FIGURE 33.18The electromagnetic spectrum. Visible light has wavelengths between 390 and 750 nm.
Vision is based on the absorption of light by photoreceptor cells in the eye. These cells are sensitive to light in a narrow region of the electromagnetic spectrum, the region with wavelengths between 390 and 750 nm (Figure 33.18). Vertebrates have two kinds of photoreceptor cells, called rods and cones because of their distinctive shapes. Cones function in bright light and are responsible for color vision, whereas rods function in dim light but do not perceive color. A human retina contains about 3 million cones and 100 million rods. Remarkably, a rod cell can respond to a single photon, and the brain requires fewer than 10 such responses to register the sensation of a flash of light.
Rhodopsin, a specialized 7TM receptor, absorbs visible light
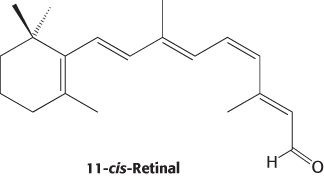
Rods are slender, elongated structures; the outer segment is specialized for photoreception (Figure 33.19). It contains a stack of about 1000 discs, which are membrane-enclosed sacs densely packed with photoreceptor molecules. The photosensitive molecule is often called a visual pigment because it is highly colored owing to its ability to absorb light. The photoreceptor molecule in rods is rhodopsin (Section 14.1), which consists of the protein opsin linked to 11-cis-retinal, a prosthetic group.
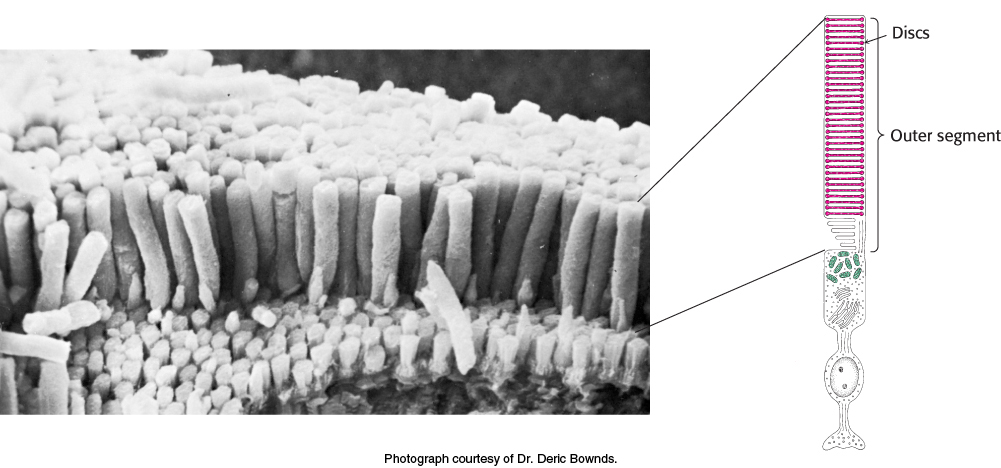
FIGURE 33.19The rod cell. (Left) Scanning electron micrograph of retinal rod cells. (Right) Schematic representation of a rod cell.
Rhodopsin absorbs light very efficiently in the middle of the visible spectrum, its absorption being centered on 500 nm, which nicely matches the solar output (Figure 33.20). A rhodopsin molecule will absorb a high percentage of the photons of the correct wavelength that strike it, as indicated by the extinction coefficient of 40,000 M −1cm−1 at 500 nm. The extinction coefficient for rhodopsin is more than an order of magnitude greater than that for tryptophan, the most efficient absorber in proteins that lack prosthetic groups.
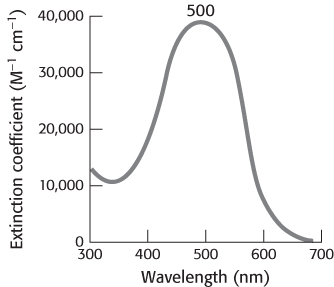
FIGURE 33.20Rhodopsin absorption spectrum. Almost all photons with wavelengths near 500 nm that strike a rhodopsin molecule are absorbed.
Opsin, the protein component of rhodopsin, is a member of the 7TM-receptor family. Indeed, rhodopsin was the first member of this family to be purified, its gene was the first to be cloned and sequenced, and its three-dimensional structure was the first to be determined. The color of rhodopsin and its responsiveness to light depend on the presence of the light-absorbing group (chromophore) 11-cis-retinal. This compound is a powerful absorber of light because it is a polyene; its six alternating single and double bonds constitute a long, unsaturated electron network. Recall that alternating single and double bonds account for the chromophoric properties of chlorophyll (Section 19.2). The aldehyde group of 11-cis-retinal forms a Schiff base (Figure 33.21) with the ε-amino group of lysine residue 296, which lies in the center of the seventh transmembrane helix. Free retinal absorbs maximally at 370 nm, and its unprotonated Schiff-base adduct absorbs at 380 nm, whereas the protonated Schiff base absorbs at 440 nm or longer wavelengths. Thus, the 500-nm absorption maximum for rhodopsin strongly suggests that the Schiff base is protonated; additional interactions with opsin shift the absorption maximum farther toward the red. The positive charge of the protonated Schiff base is compensated by the negative charge of glutamate 113 located in helix 2; the glutamate residue closely approaches the lysine–retinal linkage in the three-dimensional structure of rhodopsin.
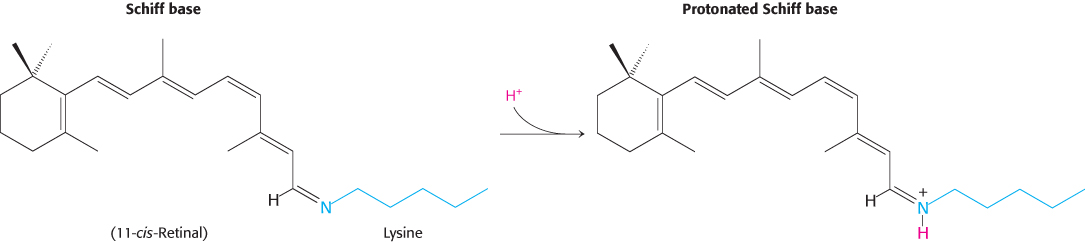
FIGURE 33.21Retinal–lysine linkage. Retinal is linked to lysine 296 in opsin by a Schiff-base linkage. In the resting state of rhodopsin, this Schiff base is protonated.
Light absorption induces a specific isomerization of bound 11-cis-retinal
How does the absorption of light by the retinal Schiff base generate a signal? George Wald and his coworkers discovered that light absorption results in the isomerization of the 11-cis-retinal group of rhodopsin to its all-trans form (Figure 33.22). This isomerization causes the Schiff-base nitrogen atom to move approximately 5 Å, assuming that the cyclohexane ring of the retinal group remains fixed. In essence, the light energy of a photon is converted into atomic motion. The change in atomic positions, like the binding of a ligand to other 7TM receptors, sets in train a series of events that lead to the closing of ion channels and the generation of a nerve impulse.
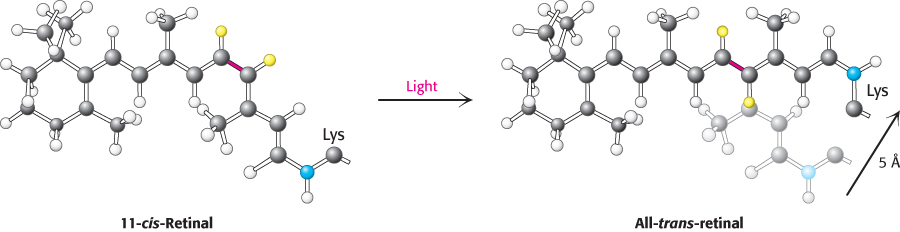
FIGURE 33.22Atomic motion in retinal. The Schiff-base nitrogen atom moves 5 Å as a consequence of the light-induced isomerization of 11-cis-retinal to all-trans-retinal by rotation about the bond shown in red.
The isomerization of the retinal Schiff base takes place within a few picoseconds of a photon being absorbed. The initial product, termed bathorhodopsin, contains a strained all-trans-retinal group. Within approximately 1 ms, this intermediate is converted through several additional intermediates into metarhodopsin II. In metarhodopsin II, the Schiff base is deprotonated and the opsin protein has undergone significant reorganization.
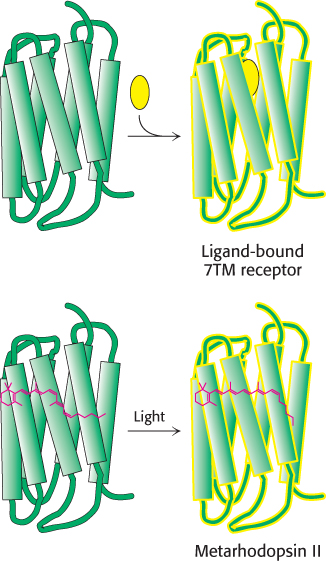
FIGURE 33.23Analogous 7TM receptors. The conversion of rhodopsin into metarhodopsin II activates a signal-transduction pathway analogously to the activation induced by the binding of other 7TM receptors to appropriate ligands.
Metarhodopsin II (also referred to as R*) is analogous to the ligand-bound state of 7TM receptors such as the β2-adrenergic receptor (Section 14.1) and the odorant and tastant receptors discussed previously (Figure 33.23). Like these receptors, this form of rhodopsin activates a heterotrimeric G protein that propagates the signal. The G protein associated with rhodopsin is called transducin. Metarhodopsin II triggers the exchange of GDP for GTP by the α subunit of transducin (Figure 33.24). On the binding of GTP, the βγ subunits of transducin are released and the α subunit switches on a cGMP phosphodiesterase by binding to an inhibitory subunit and removing it. The activated phosphodiesterase is a potent enzyme that rapidly hydrolyzes cGMP to GMP. The reduction in cGMP concentration causes cGMP-gated ion channels to close, leading to the hyperpolarization of the membrane and neuronal signaling. At each step in this process, the initial signal—the absorption of a single photon—is amplified so that it leads to sufficient membrane hyperpolarization to result in signaling.
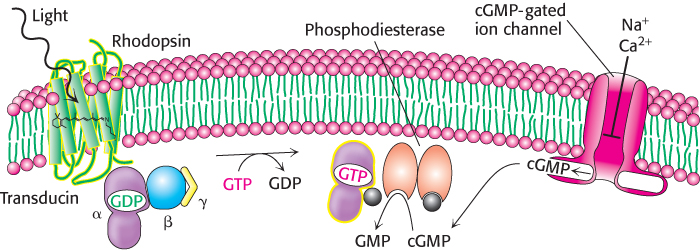
FIGURE 33.24Visual signal transduction. The light-induced activation of rhodopsin leads to the hydrolysis of cGMP, which in turn leads to ion-channel closing and the initiation of an action potential.
Light-induced lowering of the calcium level coordinates recovery
As we have seen, the visual system responds to changes in light and color within a few milliseconds, quickly enough that we are able to perceive continuous motion at nearly 1000 frames per second. To achieve a rapid response, the signal must also be terminated rapidly and the system must be returned to its initial state. First, activated rhodopsin must be blocked from continuing to activate transducin. Rhodopsin kinase catalyzes the phosphorylation of the carboxyl terminus of R* at multiple serine and threonine residues. Arrestin, an inhibitory protein then binds phosphorylated R* and prevents additional interaction with transducin.
Second, the α subunit of transducin must be returned to its inactive state to prevent further signaling. Like other G proteins, the α subunit possesses built-in GTPase activity that hydrolyzes bound GTP to GDP. Hydrolysis takes place in less than a second when transducin is bound to the phosphodiesterase. The GDP form of transducin then leaves the phosphodiesterase and reassociates with the βγ subunits, and the phosphodiesterase returns to its inactive state. Third, the level of cGMP must be raised to reopen the cGMP-gated ion channels. The action of guanylate cyclase accomplishes this third step by synthesizing cGMP from GTP.
Calcium ion plays an essential role in controlling guanylate cyclase because it markedly inhibits the activity of the enzyme. In the dark, Ca2+ as well as Na+ enter the rod outer segment through the cGMP-gated channels. Calcium ion influx is balanced by its efflux through an exchanger, a transport system that uses the thermodynamically favorable flow of four Na+ ions into the cell and one K+ ion out of the cell to extrude one Ca2+ ion. After illumination, the entry of Ca2+ through the cGMP-gated channels stops, but its export through the exchanger continues. Thus, the cytoplasmic Ca2+ level drops from 500 nM to 50 nM after illumination. This drop markedly stimulates guanylate cyclase, rapidly restoring the concentration of cGMP to reopen the cGMP-gated channels.
By controlling the rate of cGMP synthesis, Ca2+ levels govern the speed with which the system is restored to its initial state.
Color vision is mediated by three cone receptors that are homologs of rhodopsin
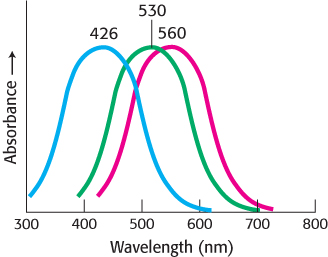
FIGURE 33.25Cone-pigment absorption spectra. The absorption spectra of the cone visual pigment responsible for color vision.
Cone cells, like rod cells, contain visual pigments. Like rhodopsin, these photoreceptor proteins are members of the 7TM-receptor family and use 11-cis-retinal as their chromophore. In human cone cells, there are three distinct photoreceptor proteins with absorption maxima at 426, 530, and ~560 nm (Figure 33.25). These absorbances correspond to the blue, green, and yellow-green regions of the spectrum. Recall that the absorption maximum for rhodopsin is 500 nm. The photoreceptor protein with its absorption maximum near 560 nm absorbs red light (with wavelength >620 nm) whereas the other two photoreceptors do not. For simplicity, we shall refer to the three proteins as the blue, green, and red photoreceptors.
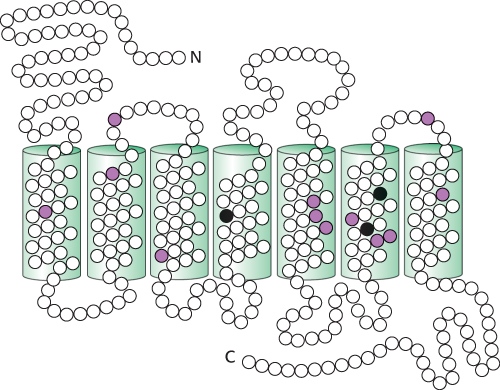
FIGURE 33.26Comparison of the amino acid sequences of the green and red photoreceptors. Open circles correspond to identical residues, whereas colored circles mark residues that are different. The differences in the three black positions are responsible for most of the difference in their absorption spectra.
The amino acid sequences of the cone photoreceptors have been compared with one another and with rhodopsin. The result is striking. Each of the cone photoreceptors is approximately 40% identical in sequence with rhodopsin. Similarly, the blue photoreceptor is 40% identical with each of the green and red photoreceptors. The green and red photoreceptors, however, are >95% identical with each other, differing in only 15 of 364 positions (Figure 33.26).
 These observations are sources of insight into photoreceptor evolution. First, the green and red photoreceptors are clearly products of a recent evolutionary event (Figure 33.27). The green and red pigments appear to have diverged in the primate lineage approximately 35 million years ago. Mammals, such as dogs and mice, that diverged from primates earlier have only two cone photoreceptors, blue and green. They are not sensitive to light as far toward the infrared region as we are, and they do not discriminate colors as well. In contrast, birds such as chickens have a total of six pigments: rhodopsin, four cone pigments, and a pineal visual pigment called pinopsin. Birds have highly acute color perception.
These observations are sources of insight into photoreceptor evolution. First, the green and red photoreceptors are clearly products of a recent evolutionary event (Figure 33.27). The green and red pigments appear to have diverged in the primate lineage approximately 35 million years ago. Mammals, such as dogs and mice, that diverged from primates earlier have only two cone photoreceptors, blue and green. They are not sensitive to light as far toward the infrared region as we are, and they do not discriminate colors as well. In contrast, birds such as chickens have a total of six pigments: rhodopsin, four cone pigments, and a pineal visual pigment called pinopsin. Birds have highly acute color perception.
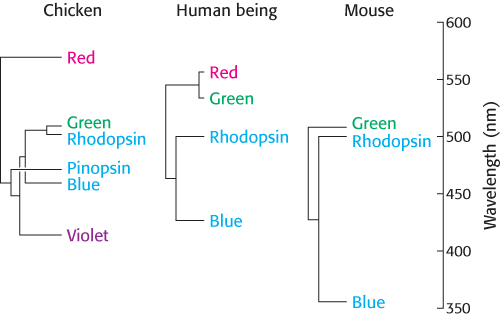
FIGURE 33.27Evolutionary relationships among visual pigments. Visual pigments have evolved by gene duplication along different branches of the animal evolutionary tree. The branch lengths of the “trees” correspond to the percentage of amino acid divergence.
[Information from J. Nathans, Neuron 24:299–312, 1999; by permission of Cell Press.]
Second, the high level of similarity between the green and the red pigments has made the identification of the specific amino acid residues responsible for spectral tuning possible. Three residues (at positions 180, 277, and 285) are responsible for most of the difference between the green and the red pigments. In the green pigment, these residues are alanine, phenylalanine, and alanine, respectively; in the red pigment, they are serine, tyrosine, and threonine. A hydroxyl group has been added to each amino acid in the red pigment. The hydroxyl groups can interact with the photoexcited state of retinal and lower its energy, leading to a shift toward the lower-energy (red) region of the spectrum.
Rearrangements in the genes for the green and red pigments lead to “color blindness”
Homologous recombination
The exchange of DNA segments with substantial sequence similarity at equivalent positions between chromosomes.
 The genes for the green and red pigments lie adjacent to each other on the human X chromosome. These genes are more than 98% identical in nucleotide sequence, including introns and untranslated regions as well as the protein-coding region. Regions with such high similarity are very susceptible to unequal homologous recombination.
The genes for the green and red pigments lie adjacent to each other on the human X chromosome. These genes are more than 98% identical in nucleotide sequence, including introns and untranslated regions as well as the protein-coding region. Regions with such high similarity are very susceptible to unequal homologous recombination.
Recombination can take place either between or within transcribed regions of the gene (Figure 33.28). If recombination takes place between transcribed regions, the product chromosomes will differ in the number of pigment genes that they carry. One chromosome will lose a gene and thus may lack the gene for, say, the green pigment; the other chromosome will gain a gene. Consistent with this scenario, approximately 2% of human X chromosomes carry only a single color-pigment gene, approximately 20% carry two, 50% carry three, 20% carry four, and 5% carry five or more. A person lacking the gene for the green pigment will have trouble distinguishing red and green color, characteristic of the most common form of color blindness. Approximately 5% of males have this form of color blindness. Recombination can also take place within the transcription units, resulting in genes that encode hybrids of the green and red photoreceptors. The absorption maximum of such a hybrid lies between that of the red and green pigments. A person with such hybrid genes who also lacks either a functional red- or green-pigment gene does not discriminate color well.
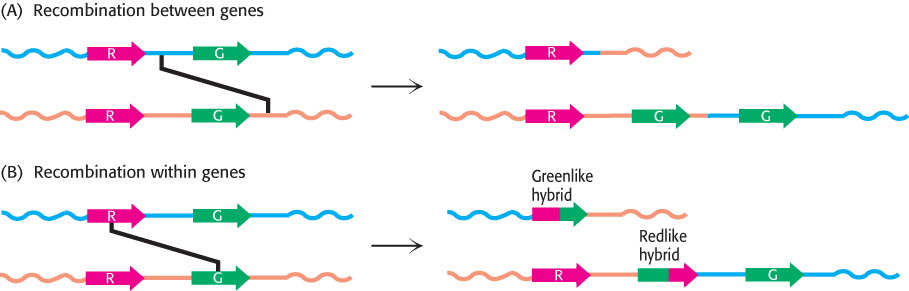
FIGURE 33.28Recombination pathways leading to color blindness. Rearrangements in the course of DNA replication may lead to (A) the loss of visual pigment genes or (B) the formation of hybrid pigment genes that encode photoreceptors with anomalous absorption spectra. Because the amino acids most important for determining absorption spectra are in the carboxyl-terminal half of each photoreceptor protein, the part of the gene that encodes this region most strongly affects the absorption characteristics of hybrid receptors.
[Information from J. Nathans, Neuron 24:299–312, 1999; by permission of Cell Press.]







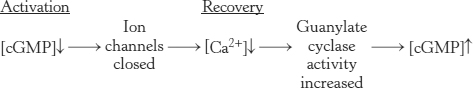


 These observations are sources of insight into photoreceptor evolution. First, the green and red photoreceptors are clearly products of a recent evolutionary event (Figure 33.27). The green and red pigments appear to have diverged in the primate lineage approximately 35 million years ago. Mammals, such as dogs and mice, that diverged from primates earlier have only two cone photoreceptors, blue and green. They are not sensitive to light as far toward the infrared region as we are, and they do not discriminate colors as well. In contrast, birds such as chickens have a total of six pigments: rhodopsin, four cone pigments, and a pineal visual pigment called pinopsin. Birds have highly acute color perception.
These observations are sources of insight into photoreceptor evolution. First, the green and red photoreceptors are clearly products of a recent evolutionary event (Figure 33.27). The green and red pigments appear to have diverged in the primate lineage approximately 35 million years ago. Mammals, such as dogs and mice, that diverged from primates earlier have only two cone photoreceptors, blue and green. They are not sensitive to light as far toward the infrared region as we are, and they do not discriminate colors as well. In contrast, birds such as chickens have a total of six pigments: rhodopsin, four cone pigments, and a pineal visual pigment called pinopsin. Birds have highly acute color perception.
 The genes for the green and red pigments lie adjacent to each other on the human X chromosome. These genes are more than 98% identical in nucleotide sequence, including introns and untranslated regions as well as the protein-
The genes for the green and red pigments lie adjacent to each other on the human X chromosome. These genes are more than 98% identical in nucleotide sequence, including introns and untranslated regions as well as the protein-