The Genetic Code Is Nonoverlapping
Investigators realized that the triplet codons in mRNA either overlapped with one another or were nonoverlapping. In a nonoverlapping code, each codon would be read as an independent unit; a single-nucleotide substitution in the mRNA would change only one codon, and the mutant protein would have only one amino acid change (Figure 17-11a). In a triplet code with maximal overlap, each codon would share two nucleotides with two other codons; a single-nucleotide change in the mRNA would alter three codons, and the resulting protein would contain three consecutive amino acid changes (Figure 17-11b).
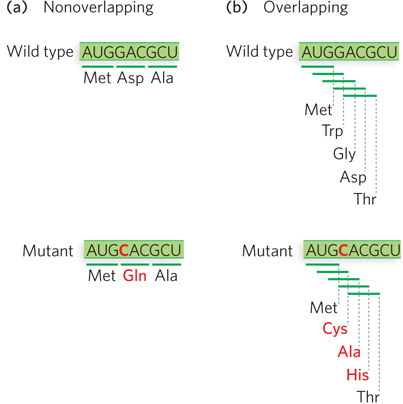
Figure 17-11: Mutation effects on nonoverlapping and overlapping codes. (a) In a nonoverlapping code, codons in the mRNA do not share nucleotides, so a single-nucleotide mutation alters only one codon, and the resulting protein has a single amino acid change. (b) In an overlapping code, some nucleotides are shared by several codons. In a triplet code with maximum overlap, a nucleotide can be shared by three codons, so a single-nucleotide mutation results in three codon changes and thus three amino acid alterations in the protein. The genetic code of all living systems is now known to be nonoverlapping.
A code with overlapping codons was ruled out experimentally by studies of mutant proteins. Most mutations result from single-base substitutions. Therefore, in a nonoverlapping code one amino acid would be changed, whereas in a maximally overlapping code three consecutive amino acids would be changed (see Figure 17-11). Independent studies of mutants of three different proteins—hemoglobin, tobacco mosaic virus protein, and tryptophan synthetase—demonstrated that the code must be nonoverlapping. In a combined total of nearly 100 different mutants of these proteins, almost all of the mutants had only one amino acid change. Thus, the genetic code is nonoverlapping, and any exceptions in the findings for mutant proteins are probably the result of double mutations.
There Are No Gaps in the Genetic Code
Investigators also understood that the codons in mRNA could be arranged one after the other, with no separation, or could be separated by one or more nucleotides acting as punctuation, like a comma. Commas between codons would prevent the deleterious effects of frameshift mutations that result from deletions or insertions of nucleotides. If there were no commas to set codons apart, a frameshift mutation would throw off the entire reading frame (Figure 17-12a). If codons were separated by a noncoding nucleotide, a frameshift mutation would result in only one amino acid change, because the next comma after the altered codon would signal the ribosome to get back on track in the correct reading frame (Figure 17-12b).

Figure 17-12: The effect of deletion mutations in codes without and with commas. (a) In a code without commas, a single-nucleotide deletion (here, deletion of a G residue) throws off the reading frame by one nucleotide. All amino acids added after this point are different from those in the wild-type protein. (b) In a code with commas separating the codons, the deletion should have minimal impact on the protein product because the next comma encountered after the deleted nucleotide would reset the ribosome to the correct reading frame. The genetic code of all living systems is now known to be without commas.
Francis Crick and Sydney Brenner performed a series of ingenious experiments in the early 1960s to determine whether the genetic code contains punctuation between codons. They studied the B gene of the T4 bacteriophage, which encodes a protein needed for the phage to grow on two different strains of E. coli. Severe mutations in the B gene restrict phage growth to one E. coli strain, but minor alterations produced near one end of the B gene are tolerated and preserve the dual-host-range phenotype. Crick and Brenner introduced mutations into the B gene chemically, using acridines as mutagens. These planar molecules intercalate into DNA and usually produce mutations through the insertion or deletion of a single base pair. The acridines were used at a concentration low enough to cause an average of one mutation per phage. The researchers tested the effects of these mutations by assaying for plaques (i.e., phage growth) on the two different host strains of E. coli.
Most mutations completely inactivated the B gene, even when the mutations mapped to the region of the gene that can tolerate minor alterations. These results suggested that not one but many amino acids were changed by the acridine-induced mutations: the mutations seemed to throw off the entire reading frame, indicating that the code has no commas to bring the ribosome back on track.
To study these mutations further, Crick and Brenner recombined the B genes from two mutant T4 phages by mixing them in a host E. coli culture, creating phages with a B gene containing two mutations instead of one. In some cases, the double-mutated gene restored the dual host range of the wild-type phage, but other pairs of mutations did not restore the dual-range phenotype. Thus, the single-mutant phages used for the recombination could be sorted into two groups, referred to as a plus or a minus group. Double mutants made by crossing two single mutants of like sign could not form a wild-type double mutant, but crossing two single mutants of opposite sign formed double mutants with the dual-host-range phenotype.
Crick and Brenner interpreted their findings as follows. The gain or loss of one base pair results in a shift in the wild-type reading frame (Figure 17-13a), thus changing all the amino acids after the point of mutation (Figure 17-13b, c). When mutants of opposite sign are combined (an insertion and a deletion), the first mutation encountered during translation causes a frameshift, but the second mutation of opposite sign restores the original reading frame (Figure 17-13d). Therefore, the only amino acid alterations that occur are located between the two mutations. Mutants of like sign do not complement one another, because they do not reestablish the correct reading frame. Overall, these results implied that codons are read in a reading frame without commas. There are no gaps between words in the genetic code.

Figure 17-13: The effects on a reading frame of combining insertion and deletion mutations. Insertion or deletion of a single nucleotide throws the ribosome into the wrong reading frame and produces a mutant protein (amino acids shown in red). (a) The wild-type protein sequence. (b) The effect of an insertion mutation. (c) The effect of a deletion mutation. (d) Combining an insertion and a deletion affects some amino acids but eventually restores the correct sequence. (e) Combining three consecutive insertion mutations (or three deletions) leaves the remaining triplets intact—evidence that a codon has three, rather than four or five, nucleotides.
The Genetic Code Is Read in Triplets
The first indication that the code is read in groups of three nucleotides came from an extension of the acridine-induced frameshift mutant studies in T4 phage. This time, Crick and Leslie Barnett combined three different mutants. They predicted that if the codons were read in sets of three nucleotides, the crossing of three B-gene mutants of T4, all with the same sign, would reestablish the reading frame and produce an active B-gene product (Figure 17-13e). Indeed, crossing three B-gene mutants of like sign (by mixing in an E. coli host culture) restored wild-type, or near wild-type, activity. The results suggested that the original reading frame was restored by either the deletion or the insertion of three nucleotides.
This was the first evidence that codons consist of three nucleotides. However, this interpretation is based on the assumption that each of the mutations in the three B-gene mutants was indeed a single-nucleotide insertion (or deletion). Without direct sequence information, it remained possible that more than one base pair was added (or removed) in one or more of the mutants. Of course, we now know that codons really do consist of three nucleotides apiece and that the genetic code is read in triplets.
Protein Synthesis Is Linear
There are many possible ways in which a chain of amino acids could be assembled on an mRNA transcript. For example, chain growth could initiate at one end of the polypeptide, either the N-terminus or the C-terminus, or it could start in the middle and grow outward in both directions. In fact, the chain could even be synthesized in random segments that were then stitched together to form the final product. In 1961, Howard Dintzis performed elegant studies on the synthesis of hemoglobin, using extracts from rabbit reticulocytes (immature red blood cells), and demonstrated that protein synthesis proceeds linearly, from the N- to the C-terminus (see the How We Know section at the end of this chapter).
Given that the direction of protein synthesis proceeds from the N-terminus to the C-terminus, determining the direction in which codons are translated became a relatively simple matter. Chemical methods of synthesizing RNAs of defined sequence had already been developed to crack the genetic code. For example, a synthetic hexanucleotide RNA should direct the synthesis of two amino acids. Thus, an RNA of sequence 5′-AAAUUU would encode Lys-Phe if the codons were read in the 5′→3′ direction, or Phe-Lys if read in the 3′→5′ direction (recall that peptides are always written in the N- to C-terminal direction). Experiments demonstrated that codons are read in the 5′→3′ direction during translation.
SECTION 17.2 SUMMARY
Single-nucleotide changes in a gene result in changes in a single amino acid residue in the protein product, demonstrating a nonoverlapping genetic code. Each codon is an independent unit, coding for a single amino acid.
The genetic code has no commas. Single-nucleotide insertion or deletion mutations result in a complete loss of activity in the mutant gene; these mutations throw off the reading frame from the point of mutation onward, because the code lacks any signal (comma) to reset the reading frame. Double mutations with an insertion and a deletion restore the reading frame.
Codons are composed of triplet sequences. Triple-insertion mutations and triple-deletion mutations result in active protein.
Protein synthesis is linear; it proceeds from the N-terminus to the C-terminus, and mRNA is read in the 5′→3′ direction.


