22.4 RNA INTERFERENCE
Thus far, we have been discussing mechanisms of gene regulation that involve regulatory proteins. These proteins bind to DNA or RNA targets and bring about changes in the efficiency of transcription or translation in response to various stimuli. However, an entirely distinct mode of gene regulation became apparent in experiments initially carried out to examine petal color in petunias (Figure 22-14) and in experiments on nematodes. These studies revealed the existence of a regulatory mechanism involving small RNA molecules—RNA interference (RNAi). Researchers discovered that when exogenous RNA was introduced into a eukaryotic cell, either experimentally or naturally, by infection with an RNA virus, the RNA was processed into 21- to 27-nucleotide species known as short interfering RNAs (siRNAs); these mediated the silencing of certain genes. Although RNAi by siRNAs was the first RNA-mediated gene silencing pathway to be discovered, researchers soon found that silencing can also occur in pathways involving small RNAs called microRNAs (miRNAs), encoded by the cells themselves. The two gene silencing mechanisms are very similar—siRNAs make use of cellular machinery associated with the endogenous miRNAs—but have some important differences. In both cases, though, these small RNAs function by base pairing with mRNAs, often in the 3′UTR, which results in mRNA degradation or translation inhibition. In some cases they can also repress transcription of the targeted mRNA.
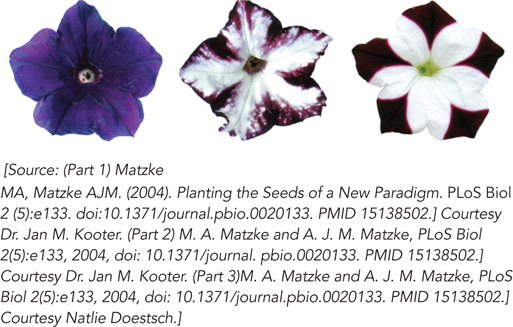
Figure 22-14: RNAi pathway effects: petal color selection in petunias. In these petunia flowers, genes for pigmentation are silenced by RNAi. The flower on the left is the wild type; the other two flowers are from transgenic plants with inserted genes that produce siRNAs complementary in sequence to an endogenous gene required for the development of flower color. This leads to the unpigmented, white areas of the petals.
RNA interference and related pathways control developmental timing in some organisms. They are also used as a mechanism to protect against invading RNA viruses (especially important in plants, which lack an immune system) and to control the activity of transposons. Small RNA molecules also play a critical, although still undefined, role in the formation of heterochromatin. In addition, RNAi has become a powerful tool for molecular biologists and has attracted attention as a potential therapeutic approach. In 2014, close to 150 trials of RNAi-based therapies were in progress. For example, an RNAi-based therapy has been developed for a rare disease called transthyretin-mediated amyloidosis, which is caused by a mutant liver protein that circulates in the blood and causes nerve damage, and even death. In an early trial, the new therapy reduced levels of the protein by as much as 96%.
Eukaryotic MicroRNAs Target mRNAs for Gene Silencing

Craig Mello

Andrew Fire
In the 1980s and 1990s, numerous experiments were under way, on a variety of organisms, to use DNA or RNA oligonucleotides antisense (complementary) to an mRNA to block protein expression. The hypothesis was that base pairing between the antisense oligonucleotide and the target mRNA would prevent recognition by the translation machinery, or lead to degradation of the hybrid complex, or both. Experiments in plants, however, showed that many transgenic plants containing an artificial gene encoding an antisense RNA failed to suppress expression of the corresponding endogenous gene. Furthermore, in both plants and nematodes, “control” experiments in which the sense strand, rather than the antisense strand, of RNA was introduced into cells often showed just as much suppression of the targeted gene as did experiments using the antisense strand. Careful analysis of these phenomena in the nematode system by Craig Mello and Andrew Fire revealed a fascinating explanation for these puzzling observations. The observed RNAi in C. elegans resulted from the presence of small amounts of double-stranded RNA that contaminated the preparations of sense or antisense RNA injected into the worms. Researchers subsequently demonstrated in plants, worms, and other eukaryotes that the double-stranded siRNAs were much more efficient at inducing gene silencing than were single-stranded sense or antisense RNAs (Figure 22-15). Mello and Fire shared the 2006 Nobel Prize in Medicine or Physiology for this discovery.
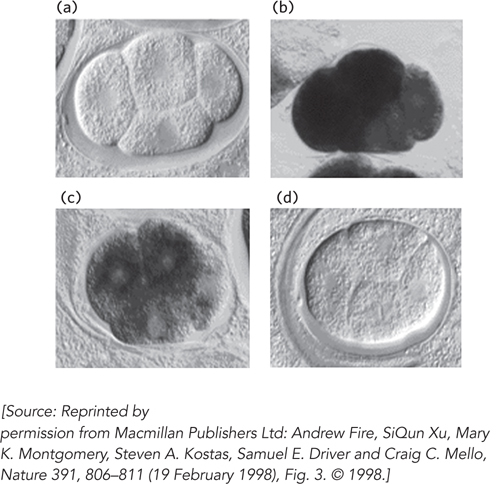
Figure 22-15: Silencing of gene expression by RNAi in a nematode. Mex-3 protein is a regulator that is expressed and functions early in nematode development. Four early embryos are shown at identical stages of development, with in situ hybridization used to detect the presence of mex-3 transcripts. (a) No staining is seen in a control, with no hybridization probe added. (b) Staining reveals the normal pattern of mex-3 expression in the embryo. (c) Injecting an embryo with an RNA complementary (antisense) to mex-3 mRNA reduces gene expression somewhat. (d) Injecting an embryo with double-stranded RNA corresponding to mex-3 reduces gene expression dramatically.
Further experiments in plants, nematodes, fruit flies, and mammals revealed many endogenous, or naturally occurring, small RNAs—microRNAs (miRNAs)—that correspond to sequences in cellular mRNAs. In fact, hundreds of different miRNAs have been identified in higher eukaryotes (see the How We Know section at the end of this chapter). They are transcribed by Pol II, or in some cases by Pol III, as primary miRNA transcripts (pri-miRNAs) with one or more sets of internally complementary sequences that can fold to form hairpinlike structures. The pri-miRNAs are cleaved by the nuclear endonuclease Drosha, a member of the ribonuclease III family of enzymes; Drosha cleaves RNA molecules that have significant double-stranded regions adjacent to single-stranded extensions of one or both strands. Drosha cleavage of its cellular substrates typically produces shortened hairpins—60 to 70 nucleotides long—with a 5′ phosphate and a two-nucleotide 3′ overhang (Figure 22-16). These partially processed precursor miRNAs (pre-miRNAs) then bind to export receptor proteins and are transported from the nucleus to the cytoplasm for further processing.
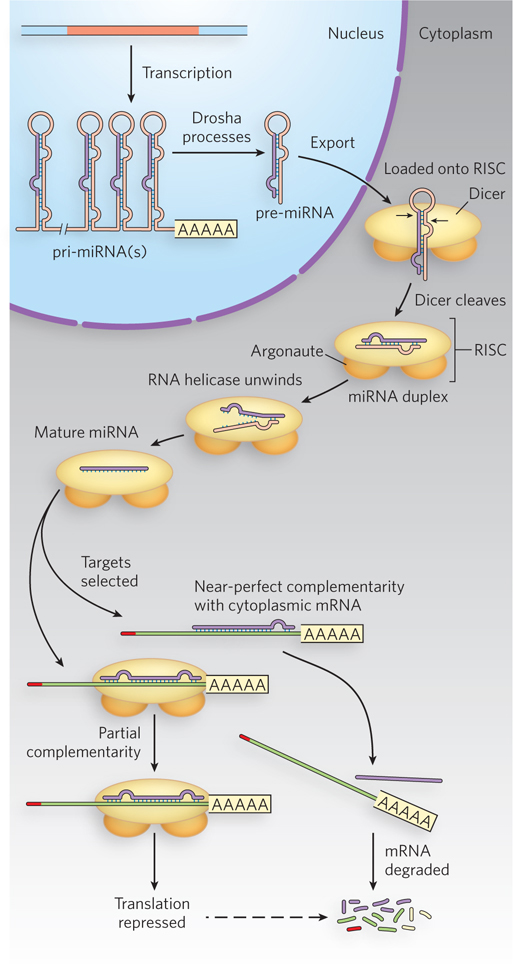
Figure 22-16: Drosha and Dicer process precursors to miRNAs. Processing of the transcribed pri-miRNA precursor by Drosha begins in the nucleus, generating a hairpin-shaped pre-miRNA that is exported to the cytoplasm. Dicer, as part of the larger RISC complex, continues the processing in the cytoplasm to generate a short double-stranded miRNA. One strand of the RNA is delivered to the target mRNA by the RISC-miRNA complex, leading to translational repression of the targeted mRNA. In plants, the targeted mRNA is often cleaved and degraded.
Once in the cytoplasm, pre-miRNAs are cleaved by Dicer, another member of the ribonuclease III family, to generate a 20 to 22 nucleotide miRNA paired with its complementary sequence. Some cells have multiple Dicer enzymes. Some of these enzymes simply cleave long double-stranded RNAs, while others are more specific for double-stranded RNAs that have a loop at one end and are of a particular length. Dicer is part of a larger complex that includes a protein from a family called Argonaute. The overall complex is called the RNA-induced silencing complex (RISC). After cleavage, the miRNA is unwound and the unneeded strand is discarded. The strand complementary to the target is delivered to particular mRNAs. Note that complementarity between the miRNA and the targeted mRNA is typically imperfect, with one or more mismatched or unmatched bases within the duplex. These mismatches usually occur two to eight nucleotides downstream from the 5′ end of the miRNA. The nucleotides at the 5′ end are called the “seed” region and must be perfectly base-paired for efficient miRNA targeting. In animals, the resulting miRNA-mRNA-protein complex triggers RISC to inhibit translation of the bound mRNA, through a process that probably blocks translation initiation. In plants, miRNAs typically induce RISC-mediated cleavage of the targeted mRNA, leading to subsequent degradation.
Although the core components of RISC are conserved across organisms, some interesting differences have been noted. For example, in fruit flies and nematodes, RISC activation requires ATP hydrolysis, whereas in mammalian systems it does not. Also, some of the proteins found in purified RISC are unique to a specific system, suggesting that RISC may be fine-tuned for function in different situations. Also of interest is that Dicer and Argonaute proteins sometimes occur in multiple isoforms, depending on the organism. Humans have just one Dicer enzyme and four Argonaute proteins, whereas nematode worms have two Dicers and many (more than 20) different Argonautes. Plants have at least eight distinct forms of Dicer. Although there may be some functional redundancy among these different protein family members, they may also play discrete roles in the implementation of RNAi-mediated control of gene expression.
Our understanding of miRNA function continues to advance, and the importance of regulation by miRNAs becomes increasingly apparent. Genes for miRNAs represent at least 4% of the genes in the human genome, or more than 1,000 genes. A given miRNA may affect the regulation of up to 400 target genes. In all, the expression of at least 60% of all human protein-encoding genes is regulated in part by miRNAs. The pattern is similar for other vertebrates.
As you may have noticed, thus far we have not mentioned yeast in our discussion of RNAi. This is because S. cerevisiae, used so prominently in the study of many other aspects of eukaryotic gene regulation, does not seem to contain any of the enzymatic machinery required for RNAi. Furthermore, attempts to induce RNAi-mediated gene silencing in budding yeast have failed. However, other single-celled eukaryotes, such as fission yeast (Schizosaccharomyces pombe) and the human pathogen Giardia intestinalis, do express Dicer and Argonaute proteins. At least in fission yeast, small endogenous RNAs are important for silencing centromeric sequences through heterochromatin formation. Why S. cerevisiae lost the capacity to use RNAi as a gene regulatory mechanism, or never acquired it, remains a fascinating question.
Short Interfering RNAs Target mRNAs for Degradation
In addition to processing pre-miRNA transcripts exported to the cytoplasm, Dicer can also recognize and cleave long double-stranded RNAs. Double-stranded RNAs can arise naturally from viral infection, or they can be introduced by experimenters. The resulting diced products, 21 to 27 bp in length (depending on the organism) with two-nucleotide 3′ overhangs, are the short interfering RNAs (siRNAs) introduced earlier, and they can regulate gene expression by mechanisms similar to those described for miRNAs. In the cytoplasm, siRNAs can form perfect or imperfect base pairings with targeted mRNA or viral RNA. In either case, these RNA duplexes are contained within the RISC protein machinery that includes Argonaute and multiple additional factors. Perfect base pairing between the siRNA and its target triggers Argonaute-catalyzed cleavage of the RNA duplex, causing eventual degradation of the targeted RNA through the normal pathways, involving 3′ deadenylation, 5′ decapping, and subsequent exonucleolytic cleavage (Figure 22-17). Imperfect pairing leads to translation repression, as described for miRNAs, and sometimes to mRNA degradation. In some cases, siRNAs can enter the nucleus and induce heterochromatin formation at the promoters of targeted genes, providing yet a third mechanism of gene silencing.
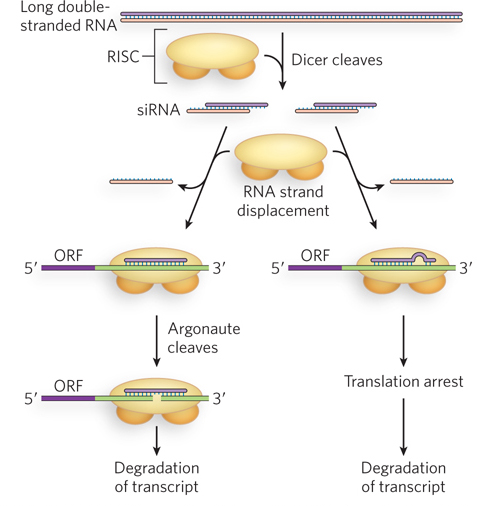
Figure 22-17: Alternative fates of siRNA-targeted mRNAs. Once the mRNA has been targeted, the degree of siRNA-mRNA base pairing leads to either Argonaute-mediated degradation of the mRNA (left) or translational repression, followed by degradation of the mRNA (right).
Much of our current understanding of siRNA function comes from experiments in nematodes, in part because of the ease and efficiency of conducting such experiments in these worms. For example, they can be fed a diet of bacteria engineered to express specified siRNA precursors, leading to the silencing of expression from siRNA-complementary mRNAs. This technique has been used in many investigations of the siRNA sequence, target site, and phenotypic consequences of siRNA-mediated gene silencing.
Another interesting aspect of RNAi in C. elegans is its extraordinary efficiency. Small amounts of siRNAs are sufficient to trigger almost complete silencing of target genes. RNA-dependent RNA polymerases have been implicated in the use of siRNAs to direct the synthesis of RNAs complementary to the targeted mRNAs, with increased numbers of siRNAs then resulting from Dicer-mediated cleavage of the new double-stranded RNAs (Figure 22-18). This amplification mechanism has not been detected in fruit flies or mammals, but does seem to occur in fission yeast (S. pombe). Furthermore, siRNA-mediated gene silencing in C. elegans can be inherited epigenetically. This means that worms in which one or more siRNAs have silenced the expression of a gene will produce offspring in which that gene remains silenced. Genetic and biochemical experiments have shown that an RNA-specific membrane channel is responsible for the transport of siRNAs from parent to fertilized oocyte, where the siRNAs are perhaps maintained by RNA-dependent RNA polymerase–mediated amplification. So far, this ability to pass siRNAs from one generation to the next has not been observed in other organisms.
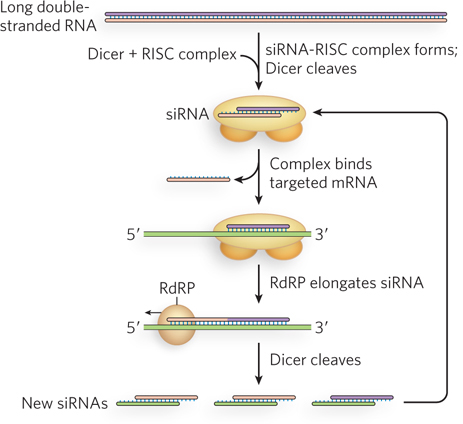
Figure 22-18: Amplification of siRNA-mediated gene regulation in nematodes by RNA-dependent RNA polymerases. The siRNAs target an mRNA by normal mechanisms. The annealed siRNA serves as a primer for RNA-dependent RNA polymerase (RdRP), which creates a longer double-stranded RNA. This is then used as a source of more siRNAs.
RNAi Pathways Regulate Viral Gene Expression

Traci M. T. Hall
Some lines of evidence suggest that RNAi originally evolved to suppress the replication of viruses and transposable elements that use double-stranded RNA as a replication intermediate. For example, plant viruses have acquired various mechanisms of suppressing the RNAi machinery in host cells. Most plant viruses have single-stranded RNA genomes that replicate through double-stranded RNA intermediates and thereby trigger RNA silencing in infected cells. On incorporation of viral-derived siRNAs into the cell’s RISC machinery, the viral RNA is specifically targeted for degradation. Genetic experiments have identified numerous viral genes that can limit this effect. In tombusviruses, one of which infects carnations, a small viral protein called p19 was found to play a role in suppressing gene silencing. Traci Hall and her colleagues found that p19 binds specifically to siRNAs, based on their length and the presence of a two-nucleotide 3′ overhang. This p19-siRNA complex cannot assemble into RISCs to direct siRNA-mediated degradation of the viral RNA (Figure 22-19).
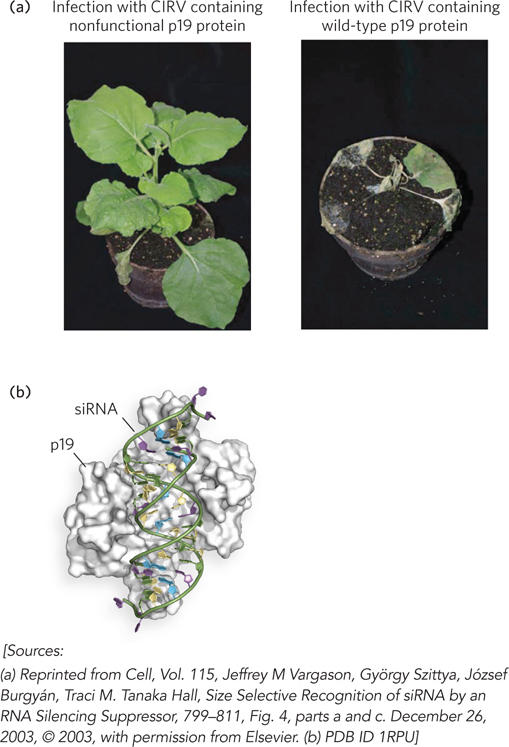
Figure 22-19: The binding of tombusvirus p19 to siRNAs and blocking of RISC assembly. (a) Plants infected with CIRV (a tombusvirus) that does not (left) or does (right) express the protein p19. (b) The structure of p19 bound to siRNA.
In an interesting twist on these plant viral siRNA-inhibitory mechanisms, experiments with hepatitis C virus (HCV) show that the virus takes advantage of a cellular miRNA to stimulate replication. HCV infects human liver cells, which express an endogenous miRNA known as miR-122. HCV is unable to replicate in cells that do not express miR-122. Moreover, HCV is unable to replicate efficiently in cultured liver cells pretreated with oligonucleotides having the antisense sequence to miR-122. Such engineered oligonucleotides, known as antagomirs, have been shown to repress viral replication in HCV-infected chimpanzees and could potentially be developed into antiviral therapeutics for humans (Highlight 22-2).
RNAi Provides a Useful Tool for Molecular Biologists
The discovery of RNA interference has an interesting and very useful practical side. If an investigator introduces into an organism a double-stranded RNA corresponding in sequence to virtually any mRNA, the Dicer endonuclease cleaves the duplex into siRNAs. These bind to the mRNA and silence it. Depending on the gene and the site(s) selected for siRNA targeting, such expression “knockdown” results in a 50% to 90% reduction in the levels of protein produced from that gene (Figure 22-20). Removal of a protein is a powerful way to investigate that protein’s cellular function. Compared with traditional genetic methods, RNAi provides a much easier way to alter gene expression for experimental or therapeutic purposes. The technique has rapidly become an important tool in ongoing efforts to study gene function, because it can disrupt functionality without creating a mutant organism.
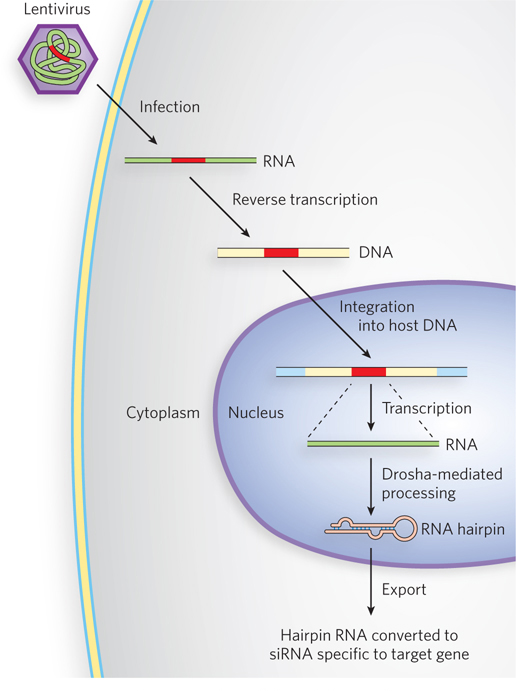
Figure 22-20: The siRNA-based knockdown of gene expression in human cells. Lentiviruses, RNA viruses that infect human cells, are used as vectors. A sequence encoding a hairpin homologous to the target gene is cloned into the lentiviral vector (red). In the host cell, the viral RNA is converted to double-stranded DNA by reverse transcriptase, and the DNA is integrated into the cellular genome. Transcription of the integrated gene produces a hairpin RNA that is converted to siRNA specific to the target gene.
In plants, virtually any gene can be effectively shut down in this way. In nematodes, simply introducing the double-stranded RNA into the worm’s diet causes effective suppression of the target gene. The procedure can be applied to human cells as well. Laboratory-produced siRNAs have already been used to block HIV and poliovirus infections in cultured human cells for a week at a time. The widespread use of this technology for medical applications has been limited by the difficulties inherent in getting the highly charged and ribonuclease-sensitive siRNAs across cellular membranes to their targets. New delivery technologies such as lipid nanoparticles and specialized polymers have begun to solve that problem. Early efforts have focused on diseases of the liver, as this organ naturally takes up a range of material from the blood—as seen in the examples of transthyretin-mediated amyloidosis and HCV infection described earlier.
HIGHLIGHT 22-2 MEDICINE: Viral Takeover Using a Cell Type–Specific miRNA

Peter Sarnow
Human tissues contain specific microRNAs that can base-pair with complementary sequences in mRNAs to trigger translational silencing and degradation. Even certain animal viruses that have RNA genomes encode miRNAs, indicating that viruses can use these small regulators during their life cycle. But how might viruses deal with, or even take advantage of, a host cell’s miRNAs during an infection?
Many viruses infect and replicate in only certain tissue types, and hepatitis C virus (HCV) is a case in point. This dangerous human pathogen selectively targets liver cells, where it can cause cirrhosis and sometimes cancer. Analysis of cultured human liver cells shows that the liver specifically expresses an miRNA called miR-122; in fact, miR-122 constitutes about 70% of the total miRNA population in liver cells. Stanford University virologist Peter Sarnow and his colleagues wondered how miR-122 regulates mRNA function in both normal and virally infected cells. They first investigated miR-122 expression levels in various tissue and cell types. Using Northern blotting (see Figure 6-32), in which miR-122 was detected using radiolabeled complementary DNA oligonucleotides, the investigators detected miR-122 in cells from intact mouse and human liver and in cultured mouse and human liver (Huh7) cells, but not in human cervical carcinoma–derived HeLa cells or even in human liver–derived HepG2 cells.
Sarnow’s research team next tried infecting these different tissues and cultured cells with HCV. Although both Huh7 and HepG2 cells are derived from human liver, HCV could replicate only in Huh7 cells. To find out whether this could be related to the presence of miR-122 in Huh7 cells, the researchers analyzed the 9,600 nucleotide HCV RNA genomic sequence for potential miR-122–binding sites that might enable a successful miRNA–target mRNA interaction. They found two sites in the noncoding regions of the viral RNA genome that are complementary to the miR-122 sequence and are conserved in many different isolates (genotypes) of HCV.
Mutation of those potential miR-122–binding sites in the viral RNA produced HCV variants that could not replicate efficiently in Huh7 cells. These mutant viruses could be “rescued” by introducing into the host Huh7 cells miR-122 variants that restored base-pair complementarity with the mutated viral RNA. Furthermore, the use of small complementary oligonucleotides to bind and sequester miR-122 in the host Huh7 cells dramatically reduced the amount of HCV RNA replication in these cells (Figure 1). Sarnow concluded that HCV requires the host cell’s miR-122 for efficient replication during infection. These findings suggest that miR-122 might be a good target for antiviral intervention.
To test this idea, Henrik Orum and his research team at Santaris Pharma in Denmark synthesized a short oligonucleotide with a sequence complementary to miR-122. To ensure that this oligonucleotide would not be rapidly destroyed in the body, they introduced chemical modifications into the sequence to block the action of degradatory enzymes. When introduced into chimpanzees chronically infected with HCV, the modified “anti–miR-122” oligonucleotide, called SPC3649, produced long-lasting suppression of HCV replication, with no evidence of viral resistance or side effects in the treated animals. This prolonged protection afforded by SPC3649 hints at a new antiviral strategy that takes aim at the underlying mechanism used by HCV to take over human liver cells.
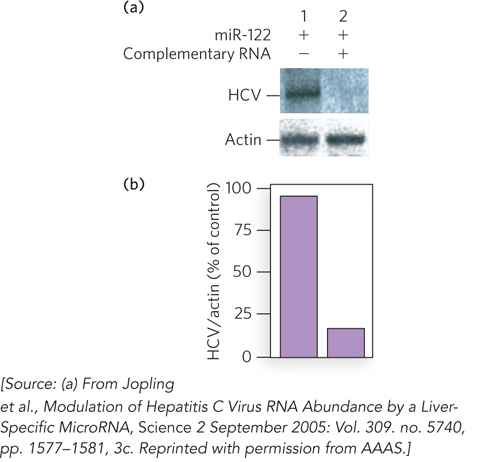
FIGURE 1 Hepatitis C virus replication is reduced in the absence of miR-122 RNA in the host cell. (a) When miR-122 was expressed in a cell harboring HCV, Northern blots showed production of HCV RNAs (lane 1). When an RNA complementary to miR-122 was also present, to sequester miR-122, HCV RNA production declined markedly (lane 2). Northern blots for a housekeeping gene that encodes actin, used as a control, are also shown. (b) Quantification of the results of the same experiments.
RNAs Regulate a Wide Range of Cellular Processes
An idea of the complexity and abundance of eukaryotic RNAs has begun to take shape, facilitated by new initiatives such as the ENCODE (Encyclopedia of DNA elements) project. Beginning with a pilot project in the early 2000s, and continuing as a much larger effort since 2007, ENCODE seeks to identify and map all functional elements in the human genome. A functional element is defined as any protein- or RNA-coding gene, transcription factor binding site, chromatin structure, or DNA methylation site. New information on RNA has been a highlight of this project. As reported in a 2012 update, more than 62% of the base pairs in the human genome are found in transcribed RNA molecules greater than 200 nucleotides in length. Regulation is a common theme, and the list of RNA-regulated processes keeps expanding. The list of small RNAs now includes the snRNAs involved in spliceosome function and the snoRNAs involved in rRNA processing (both discussed in Chapter 16), miRNAs (Chapter 16 and this chapter), RNAs that guide telomerase function (Chapter 11), RNAs involved in some types of RNA editing (Chapter 16), and many more. We cover two additional classes here.
A class of gene in multicellular eukaryotes called piwi (the name derives from a Drosophila gene called P-element induced wimpy testis) encodes regulatory proteins involved in maintaining incomplete differentiation and stable cell division in germ-line and stem cells. PIWI proteins represent a clade within a larger family that includes the Argonaute proteins involved in miRNA maturation and function. A class of short (21 to 35 nucleotides) RNAs called PIWI-interacting RNAs (piwi RNAs, or piRNAs) has been found, which bind to PIWI proteins and guide them to their RNA targets by base-pairing interactions. The piRNAs are processed from much longer, single-stranded transcripts derived from clusters of genes encoding piRNA precursors. Details of the biogenesis of piRNAs are still being worked out. In the germ line, piRNAs seem to play a variety of roles. These include the silencing of transposons (see Chapter 14) so that their movements around the genome, and the damage this would cause, are limited. The piRNAs also direct the placement of some epigenetic markers (particularly histone modifications and DNA methylation) at a variety of locations in the genome. During germ-line processes such as spermatogenesis, PIWI proteins and piRNAs trigger the opening of heterochromatin so that particular genes can be expressed, followed by piRNA-mediated reestablishment of the heterochromatin state. In mouse testes, there is a burst of piRNA expression in which piRNA production from more than 100 genes is increased about 6,000-fold during the pachytene phase of meiosis. The piRNA/PIWI complexes carry out their functions by interacting with RNAs that are being transcribed (thus, for example, preventing translation of transposase mRNAs and perhaps triggering their degradation) and recruiting other proteins and enzymes to those locations (e.g., to modify histones or methylate DNA).
The broader screen of human genomic sequences has also revealed the presence of long noncoding RNAs (lncRNAs). These are defined as RNAs longer than 200 nucleotides that do not possess a significant open reading frame to encode a protein. Some of these are transcribed from long introns in protein-coding genes. Others are antisense transcripts, derived from the nontemplate (coding) DNA strand and often overlapping with the protein-coding exon sequences. Still others are intergenic, expressed from the large genomic regions between protein-coding genes. Some of these RNAs undergo splicing and even alternative splicing. Most of the lncRNAs analyzed thus far are found to play some role in differentiation and development. In some cases, elimination of an lncRNA results in a visible developmental defect; in others, the effects are subtle. In mice, for example, an lncRNA called BC1 is derived from certain retrotransposons and is expressed at high levels in the brain. Elimination of this lncRNA does not result in physical deformity, but it does result in reduced environmental exploration and increased anxiety—behaviors likely to reduce survival in the wild. The investigation of RNA regulatory mechanisms is likely to be a major focus of molecular biology for decades to come.
SECTION 22.4 SUMMARY
RNA interference (RNAi) silences gene expression by means of short interfering RNAs (siRNAs), using endogenous gene silencing pathways based on genome-encoded microRNAs (miRNAs).
Endogenous to the cell, miRNAs are encoded by many eukaryotic genomes and cause translational inhibition of the mRNAs to which they bind. Introduced by viral infection or experimentally, exogenous siRNAs use cellular mechanisms associated with miRNAs to silence the expression of particular genes, by suppressing translation or causing mRNA degradation.
Hundreds of different miRNAs in higher eukaryotes are synthesized by Pol II or Pol III as primary miRNA transcripts (pri-miRNAs), about 70 to 90 nucleotides long. After initial processing in the nucleus, miRNAs are exported to the cytoplasm, where further processing leads to assembly into RNA-induced silencing complexes (RISCs). In contrast, siRNAs are produced entirely in the cytoplasm; like miRNAs, they assemble into RISCs.
RISCs facilitate base pairing between the small RNA and its complementary sequence in an mRNA, triggering translational arrest (when the pairing is imperfect) or mRNA degradation (when the sequences are perfectly paired).
RNAi probably evolved as a mechanism to suppress infection by RNA viruses. Molecular biologists and clinicians use RNAi to alter gene expression for experimental and therapeutic purposes.
RNAs with regulatory functions are common in eukaryotic genomes and affect a wide range of processes.











