12.2 Lessons from Yeast: The GAL System
To make use of extracellular galactose, yeast imports the sugar and converts it into a form of glucose that can be metabolized. Several genes—

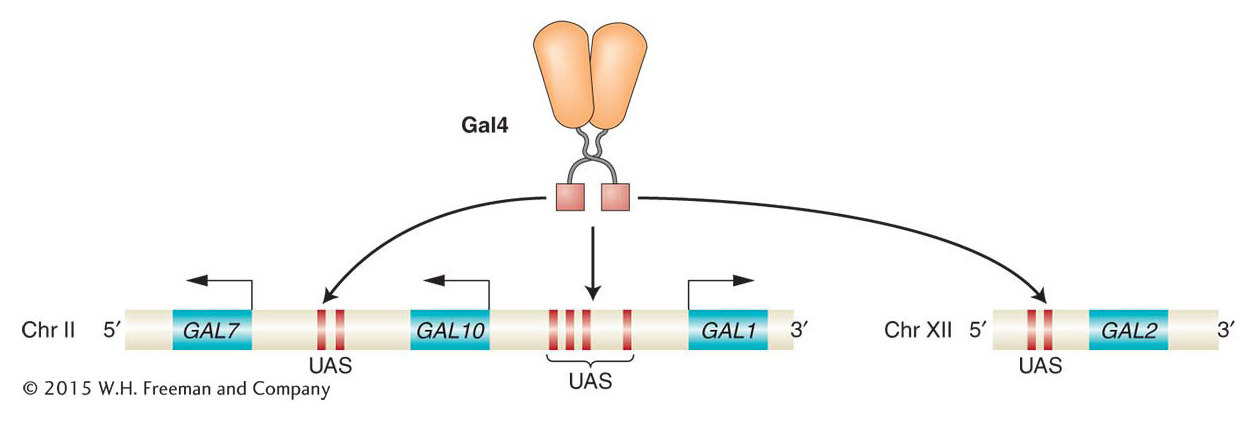
The key regulator of GAL gene expression is the Gal4 protein, a sequence-
Gal4 regulates multiple genes through upstream activation sequences
In the presence of galactose, the expression levels of the GAL1, GAL2, GAL7, and GAL10 genes are 1000-
 Yeast
Yeast
Saccharomyces cerevisiae, or budding yeast, has emerged in recent years as the premier eukaryotic genetic system. Humans have grown yeast for centuries because it is an essential component of beer, bread, and wine. Yeast has many features that make it an ideal model organism. As a unicellular eukaryote, it can be grown on agar plates and, with yeast’s life cycle of just 90 minutes, large quantities of it can be cultured in liquid media. It has a very compact genome with only about 12 mega-
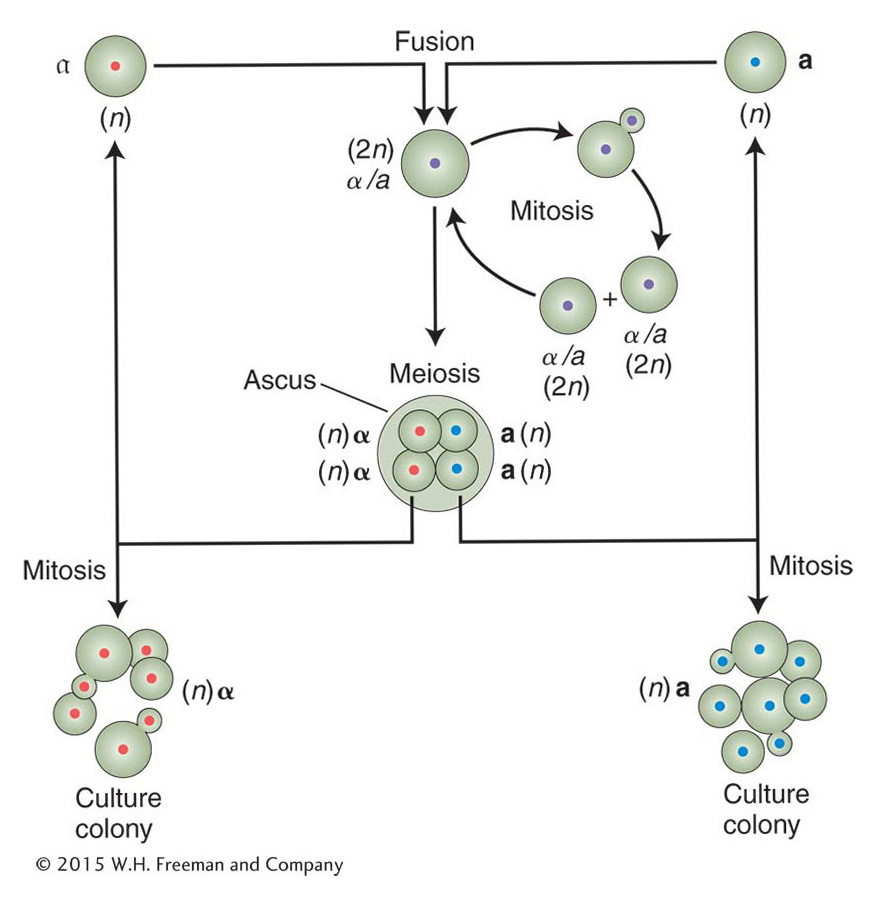
The yeast life cycle makes it very versatile for laboratory studies. Cells can be grown as either diploid or haploid. In both cases, the mother cell produces a bud containing an identical daughter cell. Diploid cells either continue to grow by budding or are induced to undergo meiosis, which produces four haploid spores held together in an ascus (also called a tetrad). Haploid spores of opposite mating type (a or α) will fuse and form a diploid. Spores of the same mating type will continue growth by budding.
Yeast has been called the E. coli of eukaryotes because of the ease of forward and reverse mutant analysis. To isolate mutants by using a forward genetic approach, haploid cells are mutagenized (with X rays, for example) and screened on plates for mutant phenotypes. This procedure is usually done by first plating cells on a rich medium on which all cells grow and by copying, or replica plating, the colonies from this master plate onto replica plates containing selective media or special growth conditions. (See also Chapter 16.) For example, temperature-
KEY CONCEPT
The binding of sequence-The Gal4 protein has separable DNA-binding and activation domains
After Gal4 is bound to the UAS element, how is gene expression induced? A distinct domain of the Gal4 protein, the activation domain, is required for regulatory activity. Thus, the Gal4 protein has at least two domains: one for DNA binding and another for activating transcription. A similar modular organization has been found to be a common feature of other DNA-
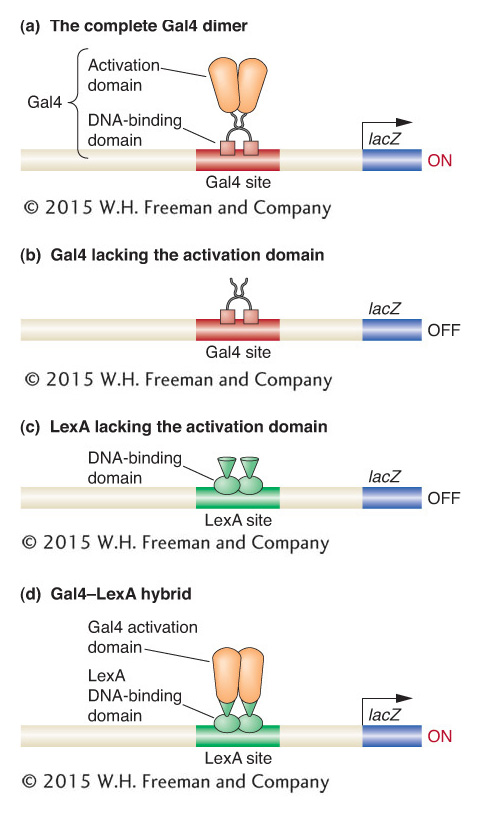
The modular organization of the Gal4 protein was demonstrated in a series of simple, elegant experiments. The strategy was to test the DNA binding and gene activation of mutant forms of the protein in which parts had been either deleted or fused to other proteins. By this means, investigators could determine whether a part of the protein was necessary for a particular function. To carry out these studies, experimenters needed a simple means of assaying the expression of the enzymes encoded by the GAL genes.
The expression of GAL genes and other targets of transcription factors is typically monitored by using a reporter gene whose level of expression is easily measured. In reporter-
Let’s see what happens when a form of the Gal4 protein lacking the activation domain is expressed in yeast. In this case, the binding sites of the UAS element are occupied, but no transcription is stimulated (Figure 12-7b). The same is true when other regulatory proteins lacking activation domains, such as the bacterial repressor LexA, are expressed in cells bearing reporter genes with their respective binding sites. The more interesting result is obtained when the activation domain of the Gal4 protein is grafted to the DNA-
KEY CONCEPT
Many eukaryotic transcriptional regulatory proteins are modular proteins, having separable domains for DNA binding, activation or repression, and interaction with other proteins.Gal4 activity is physiologically regulated
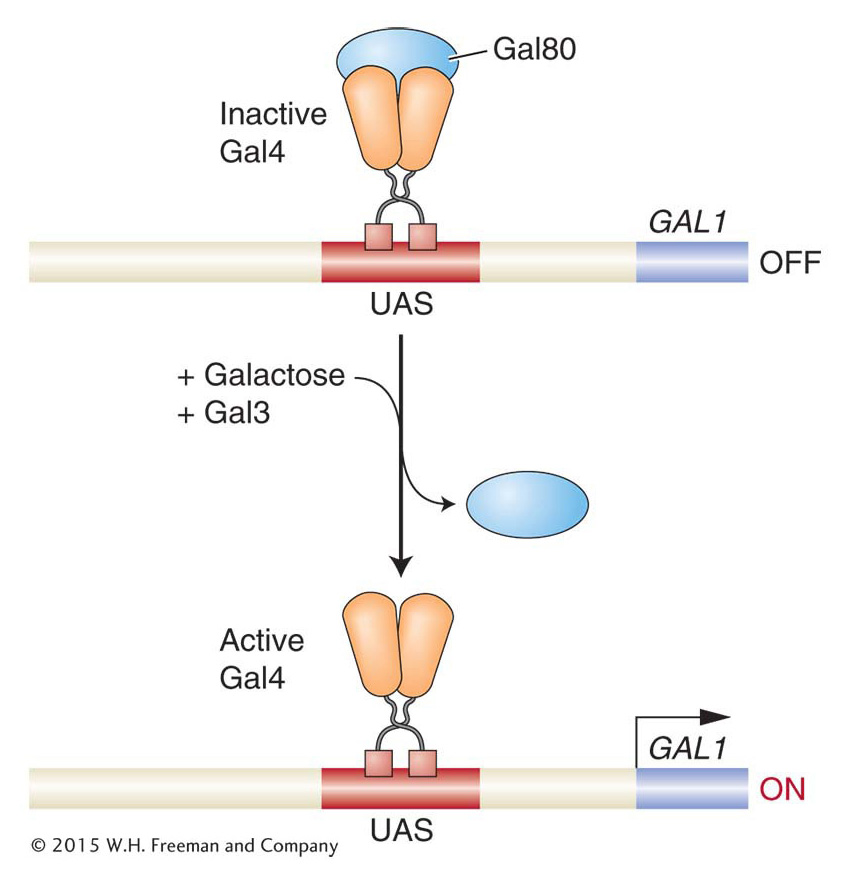
How does Gal4 become active in the presence of galactose? Key clues came from analyzing mutations in the GAL80 and GAL3 genes. In GAL80 mutants, the GAL structural genes are active even in the absence of galactose. This result suggests that the normal function of the Gal80 protein is to somehow inhibit GAL gene expression. Conversely, in GAL3 mutants, the GAL structural genes are not active in the presence of galactose, suggesting that Gal3 normally promotes expression of the GAL genes.
Extensive biochemical analyses have revealed that the Gal80 protein binds to the Gal4 protein with high affinity and directly inhibits Gal4 activity. Specifically, Gal80 binds to a region within one of the Gal4 activation domains, blocking its ability to promote the transcription of target genes. The Gal80 protein is expressed continuously, so it is always acting to repress transcription of the GAL structural genes unless stopped. The role of the Gal3 protein is to release the GAL structural genes from their repression by Gal80 when galactose is present.
Gal3 is thus both a sensor and inducer. When Gal3 binds galactose and ATP, it undergoes an allosteric change that promotes binding to Gal80, which in turn causes Gal80 to release Gal4, which is then able to interact with other transcription factors and RNA pol II to activate transcription of its target genes. Thus, Gal3, Gal80, and Gal4 are all part of a switch whose state is determined by the presence or absence of galactose (Figure 12-8). In this switch, DNA binding by the transcriptional regulator is not the physiologically regulated step (as is the case in the lac operon and bacteriophage λ); rather, the activity of the activation domain is regulated.
KEY CONCEPT
The activity of eukaryotic transcriptional regulatory proteins is often controlled by interactions with other proteins.Gal4 functions in most eukaryotes
In addition to its action in yeast cells, Gal4 has been shown to be able to activate transcription in insect cells, human cells, and many other eukaryotic species. This versatility suggests that biochemical machinery and mechanisms of gene activation are common to a broad array of eukaryotes and that features revealed in yeast are generally present in other eukaryotes and vice versa. Furthermore, because of their versatility, Gal4 and its UAS elements have become favored tools in genetic analysis for manipulating gene expression and function in a wide variety of model systems.
KEY CONCEPT
The ability of Gal4, as well as other eukaryotic regulators, to function in a variety of eukaryotes indicates that eukaryotes generally have the transcriptional regulatory machinery and mechanisms in common.Now we look at how activators and other regulatory proteins interact with the transcriptional machinery to control gene expression.
Activators recruit the transcriptional machinery
In bacteria, activators commonly stimulate transcription by interacting directly with DNA and with RNA polymerase. In eukaryotes, activators generally work indirectly. Eukaryotic activators recruit RNA polymerase II to gene promoters through two major mechanisms. First, activators can interact with subunits of the protein complexes having roles in transcription initiation and then recruit them to the promoter. Second, activators can recruit proteins that modify chromatin structure, allowing RNA polymerase II and other proteins access to the DNA. Many activators, including Gal4, have both activities. We’ll examine the recruitment of parts of the transcriptional initiation complex first.
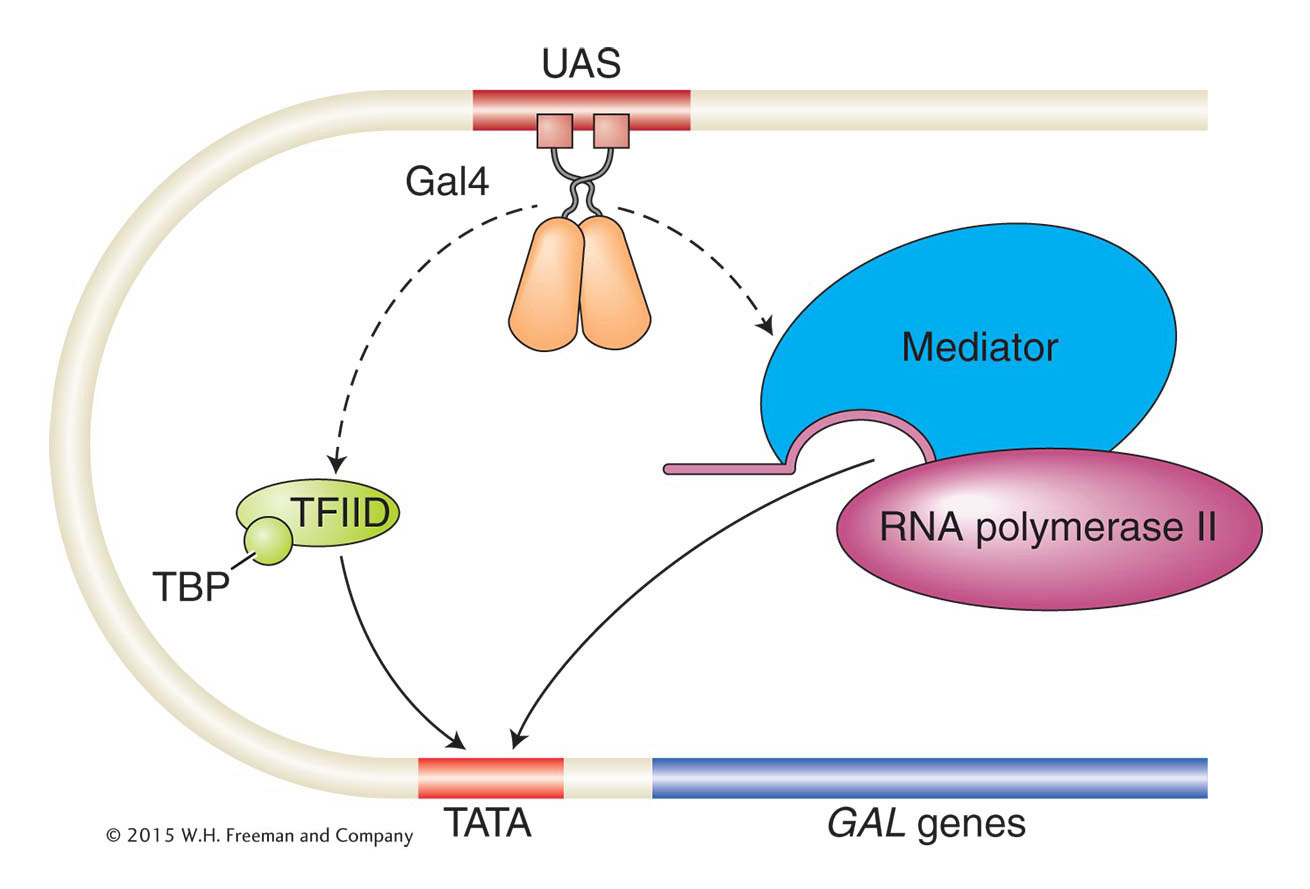
Recall from Chapter 8 that the eukaryotic transcriptional machinery contains many proteins that are parts of various subcomplexes within the transcriptional apparatus that is assembled on gene promoters. One subcomplex, transcription factor IID (TFIID), binds to the TATA box of eukaryotic promoters through the TATA-
A second way that Gal4 works to activate gene expression is by interacting with the mediator complex, a large multiprotein complex that, in turn, directly interacts with RNA polymerase II to recruit it to gene promoters. The mediator complex is an example of a co-activator, a term applied to a protein or protein complex that facilitates gene activation by a transcription factor but that itself is neither part of the transcriptional machinery nor a DNA-
The ability of transcription factors to bind to upstream DNA sequences and to interact with proteins that bind directly or indirectly to promoters helps to explain how transcription can be stimulated from more distant regulatory sequences (see Figure 12-9).
KEY CONCEPT
Eukaryotic transcriptional activators often work by recruiting parts of the transcriptional machinery to gene promoters.The control of yeast mating type: combinatorial interactions
Thus far, we have focused in this chapter on the regulation of single genes or a few genes in one pathway. In multicellular organisms, distinct cell types differ in the expression of hundreds of genes. The expression or repression of sets of genes must therefore be coordinated in the making of particular cell types. One of the best-
The yeast Saccharomyces cerevisiae can exist in any of three different cell types known as a, α, and a/α. The two cell types a and α are haploid and contain only one copy of each chromosome. The a/α cell is diploid and contains two copies of each chromosome. Although the two haploid cell types cannot be distinguished by their appearance in the microscope, they can be differentiated by a number of specific cellular characteristics, principally their mating type (see the Model Organism box). An α cell mates only with an a cell, and an a cell mates only with an α cell. An α cell secretes an oligopeptide pheromone, or sex hormone, called α factor that arrests a cells in the cell cycle. Similarly, an a cell secretes a pheromone, called a factor, that arrests α cells. Cell arrest of both participants is necessary for successful mating. The diploid a/α cell does not mate, is larger than the α and a cells, and does not respond to the mating hormones.
Genetic analysis of mutants defective in mating has shown that cell type is controlled by a single genetic locus, the mating-
How does the MAT locus control cell type? Genetic analyses of mutants that cannot mate have identified a number of structural genes that are separate from the MAT locus but whose protein products are required for mating. One group of structural genes is expressed only in the α cell type (α-specific genes), and another set is expressed only in the a cell type (a-specific genes). The MAT locus controls which of these sets of structural genes is expressed in each cell type. The MATa allele causes the structural genes of the a-type cell to be expressed, whereas the MATα allele causes the structural genes of the α-type cell to be expressed. These two alleles activate different sets of genes because they encode different regulatory proteins. In addition, a regulatory protein not encoded by the MAT locus, called MCM1, plays a key role in regulating cell type.
The simplest case is the a cell type (Figure 12-10a). The MATa locus encodes a single regulatory protein, a1. However, a1 has no effect in haploid cells, only in diploid cells. In a haploid a cell, the regulatory protein Mcm1 turns on the expression of the structural genes needed by an a cell, by binding to regulatory sequences within promoters for a-specific genes.
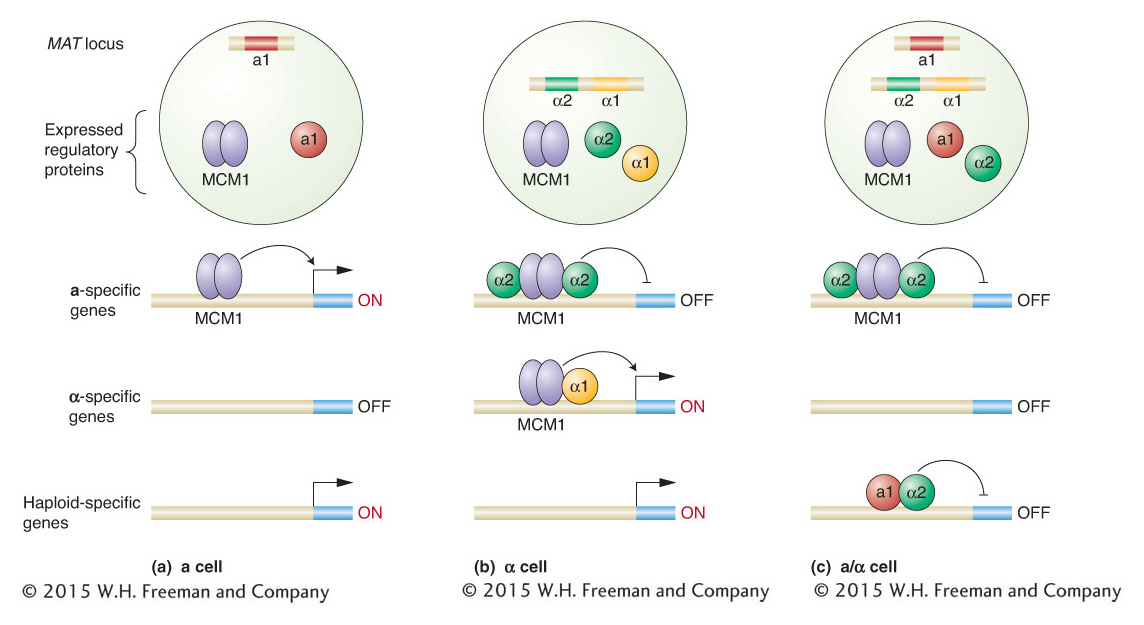
In an α cell, the α-specific structural genes must be transcribed, but, in addition, the MCM1 protein must be prevented from activating the a-specific genes. The DNA sequence of the MATα allele encodes two proteins, α1 and α2, that are produced by separate transcription units. These two proteins have different regulatory roles in the cell, as can be demonstrated by analyzing their DNA-
In a diploid yeast cell, regulatory proteins encoded by each MAT locus are expressed (Figure 12-10c). What is the result? All the structural genes involved in cell mating are shut down, as are a separate set of genes, called haploid specific, that are expressed in haploid cells but not diploid cells. How does this happen? The a1 protein encoded by MATa has a part to play at last. The a1 protein can bind to some of the α2 protein present and alter its binding specificity such that the a1–