Multiple Factors Control the Pluripotency of ES Cells
During the earliest stages of embryogenesis, as the zygote begins to divide, both the paternal and maternal DNA become demethylated (see the discussion of DNA methylation in Chapter 9). This happens in part because a key maintenance methyl transferase, Dnmt1, is transiently excluded from the nucleus and in part because demethylase enzymes actively remove or “erase” methylation marks from 5-
Page 982
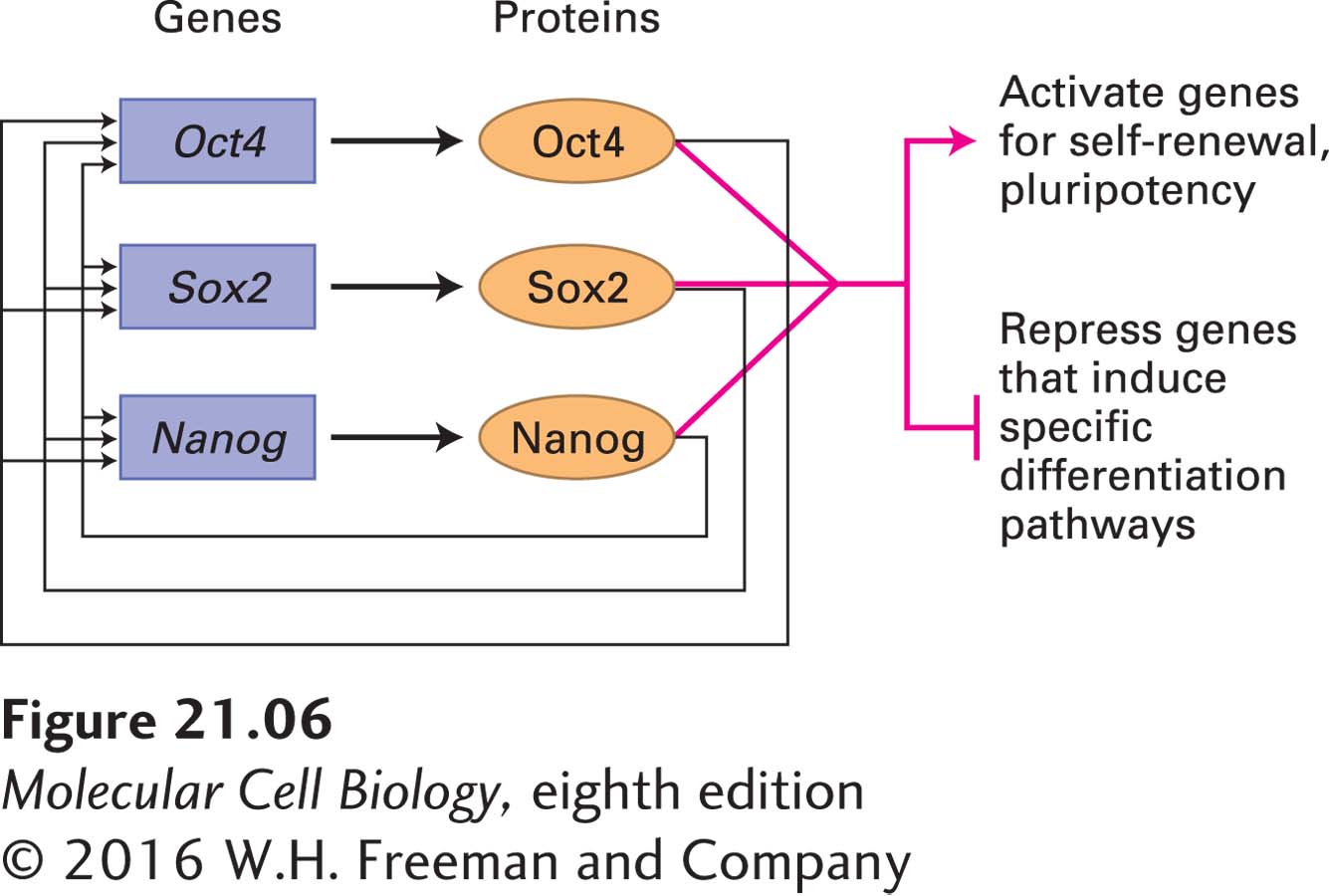
ES cell properties are also critically dependent on the action of master transcription factors produced shortly after fertilization. The transcription factors Oct4, Sox2, and Nanog have essential roles in early development and are required for the specification of ICM cells in the embryo as well as for the specification of ES cells in culture. The expression of Oct4 and Nanog is exclusive to pluripotent cells such as the cells of the ICM and cultured ES cells. Sox2 is found in pluripotent cells, but its expression is also necessary in the multipotent neural stem cells that give rise exclusively to neuronal and glial cell types (discussed in Chapter 22). Genetic studies in the mouse suggest that these three regulators have distinct roles, but may function in related pathways to maintain the developmental potential of pluripotent cells. For example, disruption of Oct4 or Sox2 results in the inappropriate differentiation of ICM and ES cells into trophectoderm. However, forced expression of Oct4 in ES cells leads to a phenotype that is similar to that caused by loss of Nanog function. Thus knowledge of the set of genes regulated by these transcription factors might reveal their essential roles during development.
The genes that are bound by these three transcription factors have been identified using chromatin immunoprecipitation experiments (see Chapter 9); each protein is found at more than a thousand chromosomal locations. The target genes encode a wide variety of proteins, including the Oct4, Nanog, and Sox2 proteins themselves, forming an autoregulatory loop in which each of these three transcription factors induces its own expression as well as that of the others (Figure 21-6). These transcription factors also bind to the transcription-
Several protein hormones are provided by feeder cells or added to culture media to prevent differentiation of ES cells. These hormones include leukemia inhibitory factor (LIF), which activates Stat3; Wnt, which activates the β-catenin transcription factor; and bone morphogenetic protein 4 (BMP4), which activates the Smad1 transcription factor (see Chapter 16). In ES cells, these three transcription factors bind at multiple genomic sites co-
Chromatin regulators that control gene transcription (see Chapter 9) are also important in ES cells. In Drosophila, Polycomb group proteins form complexes to maintain gene repression states that have been previously established by DNA-
Many other regulators play important roles in controlling gene expression and maintaining pluripotency during very early development. For example, the gene encoding the miRNA let-
Page 983
As we will see later, the possibility of using embryonic stem cells therapeutically to restore or replace damaged tissue is fueling much research on how to induce them to differentiate into specific cell types. Apart from their possible benefit in treating disease, ES cells have already proved invaluable for producing mouse mutants useful in studying a wide range of diseases, developmental mechanisms, behavior, and physiology. Using the recombinant DNA techniques described in Chapter 6, one can eliminate or modify the function of a specific gene in ES cells (see Figure 6-38). The mutated ES cells can then be employed to produce mice with a gene knockout (see Figure 6-39). Analysis of the effects of deleting or modifying a gene in this way often provides clues about the normal function of the gene and its encoded protein.