Chapter 15 Summary
Core Concepts Summary
15.1 Genetic variation describes common genetic differences (polymorphisms) among the individuals in a population at any given time.
The genotype of an organism is its genetic makeup. page 310
The phenotype of an organism is any observable characteristic of that organism, such as its appearance, physiology, or behavior. The phenotype results from a complex interplay of the genotype and the environment. page 310
A person’s genotype can be homozygous, in which he or she has two alleles of the same type (one from each parent), or heterozygous, in which he or she has a different type of allele from each parent. page 311
Genetic variation can be neutral (no effect on survival or reproduction), harmful (associated with decreased survival or reproduction), or beneficial (associated with increased survival or reproduction). page 311
15.2 Human genetic variation can be detected by DNA typing, which can uniquely identify each individual.
A variable number tandem repeat (VNTR) results from differences in the number of small-
DNA typing (DNA fingerprinting) analyzes genetic polymorphisms at multiple genes with multiple alleles. Because there are so many possible genotypes in the population, this procedure can uniquely identify an individual with high probability. page 315
A restriction fragment length polymorphism (RFLP) results from small changes in the DNA sequence between chromosomes, such as point mutations, that create or destroy restriction sites. page 316
15.3 Two common types of genetic polymorphism are single-
A SNP is a common difference in a single nucleotide. The DNA in any two human genomes differs at about 3 million SNPs. Most SNPs are not associated with any detectable effect on phenotype. page 316
CNV is a difference in the number of copies of a particular DNA sequence among chromosomes. page 318
15.4 Chromosomal variants can also occur but are usually harmful.
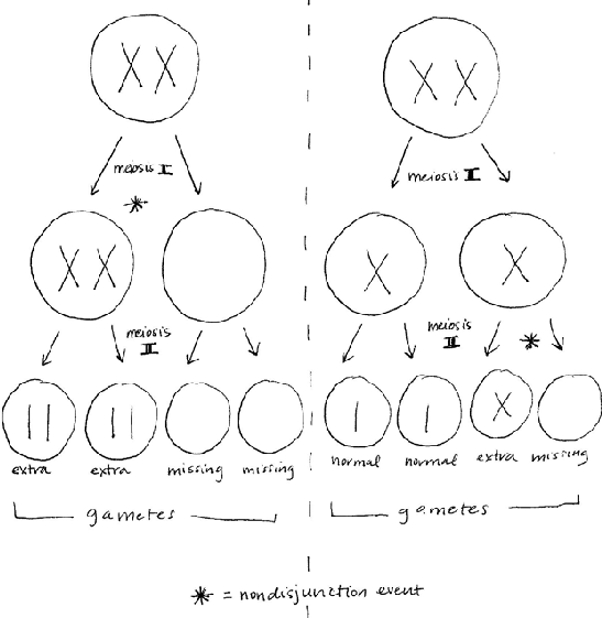
Nondisjunction is the failure of a pair of chromosomes to separate during anaphase of cell division. It can occur in mitosis or meiosis and results in daughter cells with extra or missing chromosomes. page 319
Nondisjunction in meiosis leads to gametes with extra or missing chromosomes. page 319
Most human fetuses with extra or missing chromosomes spontaneously abort, but some are viable and exhibit characteristic syndromes. page 319
Trisomy 21 (Down syndrome) is usually characterized by three copies of chromosome 21. page 320
Extra or missing sex chromosomes (X or Y) are common and result in syndromes such as Klinefelter syndrome (47, XXY) and Turner syndrome (45, X). page 322
Self-Assessment
Explain the terms “genotype” and “phenotype.” What determines a genotype? What determines a phenotype?
Self-Assessment 1 Answer
A genotype refers to the genetic composition of a cell or organism, which can differ based on the nucleotide sequence in the region(s) of interest. Although there can be many different forms (alleles) of a gene or sequence, each cell or individual normally has only two copies of each sequence, and they can either be identical to one other (homozygous) or different from one another (heterozygous). A phenotype of a cell or organism refers to its observable features, which are determined in part by its genotype, but can also be influenced by environmental factors. Some phenotypes are easily seen and vary widely among individuals, like natural hair color or height, but a phenotype may also involve less easily observable characteristics like blood type, which fall into a few distinct groups.
Describe the relationship between a genotype and a phenotype: Does the same genotype always result in the same phenotype? Must individuals with the same phenotype have the same genotype?
Self-Assessment 2 Answer
Because of environmental effects on phenotype, the same genotype does not always result in the same phenotype, and individuals with the same phenotype can have different genotypes.
With regard to mutations, what is meant by the terms “harmful,” “beneficial,” and “neutral”? Why it is sometimes an oversimplification to consider a mutation as either harmful, beneficial, or neutral?
Self-Assessment 3 Answer
Beneficial mutations are those that provide some sort of advantage to the organism in its environment, such as the Δ32 allele of the CCR5 gene that can reduce HIV infection rates in exposed individuals and delay progression to AIDS after HIV infection has occurred. The PiZ allele of the gene encoding α1AT is an example of a harmful mutation because it results in reduced levels of α1AT that, in the homozygous state, can lead to the development of emphysema due to increased breakdown of elastin in the lungs. Neutral mutations provide neither benefit nor harm to the individual, such as the AVI allele present in PTC nontasters since the inability to taste this chemical does not affect the survival or reproductive capacity of the individual. Designating a mutation as simply beneficial, harmful, or neutral does not account for the influence of the environment on the organism, which may determine how the mutation ultimately affects the individual’s ability to reproduce. For example, when an individual is heterozygous for the S allele of β-globin, he or she may have mild symptoms of blood disease (which would be considered harmful), but these individuals are also partially protected from malaria, which is highly beneficial for individuals in regions where malaria is widespread.
Describe two types of genetic polymorphism that are useful in DNA typing.
Self-Assessment 4 Answer
Differences in the copy number of tandemly repeated DNA sequences or point mutations that create or destroy restriction enzyme recognition sites are two types of differences in DNA sequences that can be used to uniquely identify an individual. Variable number of tandem repeats (VNTRs) are generated by the repetition of short stretches of DNA, one after another, in a particular region of a given chromosome. The size of the fragment generated by PCR amplification of these regions can reveal differences in the number of repeats within the region of interest. Point mutations in a DNA sequence can also introduce or disrupt a recognition site for a restriction endonuclease, so the pattern of restriction fragments that are generated by digestion of the region of interest can be used to determine whether a polymorphism exists in that region. Since each of these types of polymorphisms occurs at many different regions within the genome, the combination of the alleles at several positions can be used to distinguish the DNA of one individual from another.
Define the term “SNP” and explain why researchers are interested in detecting SNPs.
Self-Assessment 5 Answer
A SNP is a single nucleotide polymorphism, or in other words, a difference between two sequences at a single nucleotide position within a DNA sequence (point mutation). By definition, SNPs must be common enough to be found at least once in a random sample of 50 individuals, but the effect of a given SNP on phenotype can vary widely. Some SNPs are neutral, whereas others are beneficial or can increase disease risk. SNPs that correlate with disease risk or incidence are of particular interest to researchers as a potential way to identify those who are at risk for particular diseases before the onset of symptoms so that preventative treatment or early medical interventions can be initiated.
Diagram how nondisjunction in meiosis I or II can result in extra or missing chromosomes in reproductive cells (gametes).
Self-Assessment 6 Answer
Describe the consequences of an extra copy of chromosome 21 (Down syndrome).
Self-Assessment 7 Answer
Individuals with Down syndrome have three copies of chromosome 21, so they have an additional copy of the genes and elements present on this chromosome. Although the exact gene(s) responsible for this syndrome are still unknown, the increased dosage of chromosome 21 results in a group of traits that are characteristic of Down syndrome, including mild to moderate mental disability, short stature, unique facial features, and shorter life expectancy.