CONCEPT13.3 Genes Come from Various Sources and Can Be Manipulated
A major goal of molecular cloning experiments is to elucidate the functions of the DNA sequences and the proteins they encode. In this concept we look at ways that specific sequences can be identified and amplified. Millions of copies of a sequence are needed in order to study and manipulate the sequence in the laboratory. Cloned or amplified DNA can be used for various purposes, including the detection of expressed genes in specific cells and the artificial regulation of gene expression.
DNA fragments for cloning can come from several sources
The DNA fragments used in cloning procedures are obtained from a number of sources. In many cases the first step is to create a “library” of DNA fragments: a collection of clones that can be searched for the gene or genes of interest, or analyzed in other ways to learn more about the original source of the DNA fragments.
Genomic Libraries
A genomic library is a collection of DNA fragments that together comprise the genome of an organism. This is the starting point for some methods of genome sequencing. Restriction enzymes or other means, such as mechanical shearing, can be used to break chromosomes into smaller pieces (FIGURE 13.7A). Each fragment is inserted into a vector, which is then taken up by a host cell. Proliferation of a single transformed cell on a selective medium (such as for antibiotic resistance) produces a colony of recombinant cells, each of which harbors many copies of the same fragment of DNA. The colonies are grown by spreading the transformed cells over a solid culture medium in petri dishes (small circular plates), which are incubated at a suitable temperature for the host cells to grow.
RESEARCH TOOLS
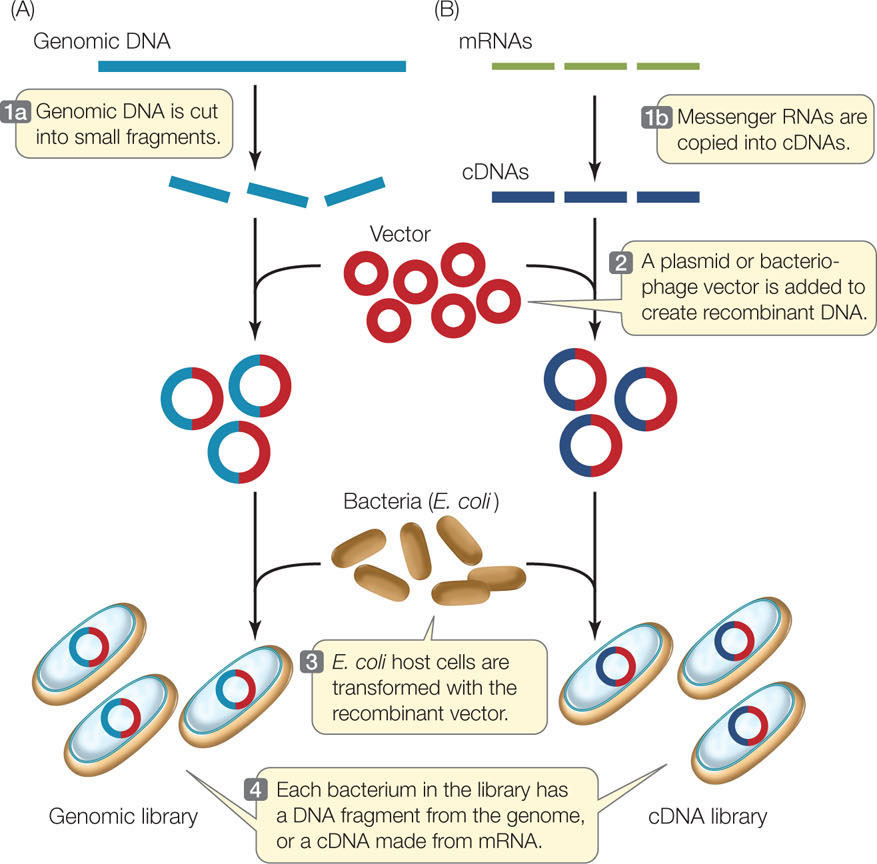
A single petri dish can hold thousands of bacterial colonies and is easily screened for the presence of a particular DNA sequence. Colonies containing that sequence are identified by DNA hybridization using a probe labeled with complementary fluorescent or radioactive nucleotides. To do this, the petri dish with its bacterial colonies is duplicated, and then the bacteria on one of the plates are treated to expose the DNA for hybridization (see Figure 10.7).
cDNA
A much smaller DNA library—one that includes only the genes transcribed in a particular tissue—can be made from complementary DNA, or cDNA (FIGURE 13.7B). This involves isolating mRNA from cells and making cDNA copies of that mRNA by complementary base pairing. The enzyme reverse transcriptase catalyzes this reaction. This collection of cDNAs from a particular tissue at a particular time is called a cDNA library, which is a “snapshot” of the transcription pattern of the cells in the sample. cDNA libraries have been invaluable for comparing gene expression in different tissues at different stages of development. For example, if cDNAs derived from developing red blood cells are examined, the globin sequences (encoding the subunits of hemoglobin) are prominent. But a cDNA library derived from hair follicles does not contain those sequences.
262
Reverse transcriptase along with PCR (see below) can be used to create and amplify a specific cDNA sequence without the need to make a library. In this case, RNA is isolated from cells and then reverse transcriptase is used to make cDNA from the RNA. Then PCR is used to amplify a specific sequence directly from the cDNA. This method, called RT-PCR, is an invaluable tool for studies of the expression of particular genes in cells and organisms.
Synthetic DNA can be made in the laboratory
In Concept 9.2 (see Figure 9.15) we described the polymerase chain reaction (PCR), a method of amplifying DNA in a test tube. PCR can begin with just a single molecule of DNA, although larger quantities [in the picogram (10−12) to microgram (10−6) range] are more often used. Any fragment of DNA can be amplified as long as appropriate primers are available. This amplified DNA can then be inserted into a plasmid to create recombinant DNA, and cloned in host cells.
The artificial synthesis of DNA by organic chemistry methods is now fully automated. Synthetic oligonucleotides (single-stranded DNA fragments of 20–40 bp) are used as primers in PCR reactions. These primers can be designed to create short new sequences at the ends of the PCR products. This might be done to create a mutation in a recombinant gene, or to add restriction enzyme sites at the ends of the PCR product to aid in ligation reactions. Longer synthetic sequences can be pieced together to construct completely artificial genes that have been designed for specific purposes. For example, a gene might be designed to be highly expressed in a particular cell type, or to encode a highly active enzyme.
LINK
Synthetic DNA was used to create a novel bacterial genome to replace the genome in a host cell, resulting in a new bacterial species; see the opening story of Chapter 4
DNA sequences can be manipulated to study cause-and-effect relationships
Mutations that occur in nature have been important in demonstrating cause-and-effect relationships in biology. However, mutations in nature are rare events. Recombinant DNA technology allows us to ask “what if” questions by creating artificial gene constructs. Because synthetic DNA can be made with any desired sequence, it can be manipulated to create specific constructs or mutations, and the resulting phenotypes can be observed when the recombinant DNA is expressed in host cells. Such techniques have revealed thousands of cause-and-effect relationships.
One example involves the auxin response element, a short sequence of DNA that binds a specific transcription factor. This element is found in the promoters of plant genes that are switched on in the presence of the plant hormone auxin (see Concept 26.2). To study the role of the auxin response element in plants, scientists made an artificial promoter containing many copies of the element, and ligated the promoter to a reporter gene. The recombinant DNA was used to transform Arabidopsis plants. When the plants were treated with auxin, the reporter gene was switched on at very high levels (higher than those produced by a wild-type auxin-responsive promoter). This experiment helped show that the presence of the auxin response element (the “cause”) results in gene expression in response to auxin (the “effect”).
Genes can be inactivated by homologous recombination
Another way to understand a gene’s function is to inactivate it so it is not transcribed and translated into a protein. An example of this approach is the use of transposon mutagenesis in experiments designed to describe the minimal genome (see Figure 12.8). In animals, these “knockout” experiments often involve homologous recombination rather than transposon mutagenesis. As we saw in Chapter 8, recombination occurs when a pair of homologous chromosomes line up during meiosis. The chromosomes sometimes break and then rejoin in such a way that segments of the two chromosomes are exchanged. A key feature of homologous recombination is that it involves an exchange of DNA between molecules with identical, or nearly identical, sequences.
263
We will focus here on the technique used for mice (FIGURE 13.8). In order to knock out (inactivate) a target gene, the normal allele of the gene is inserted into a plasmid. Restriction enzymes are then used to insert a fragment containing a reporter gene or selectable marker into the middle of the normal gene. This addition of extra DNA disrupts the gene’s coding region so that it no longer encodes a functional protein product.
RESEARCH TOOLS
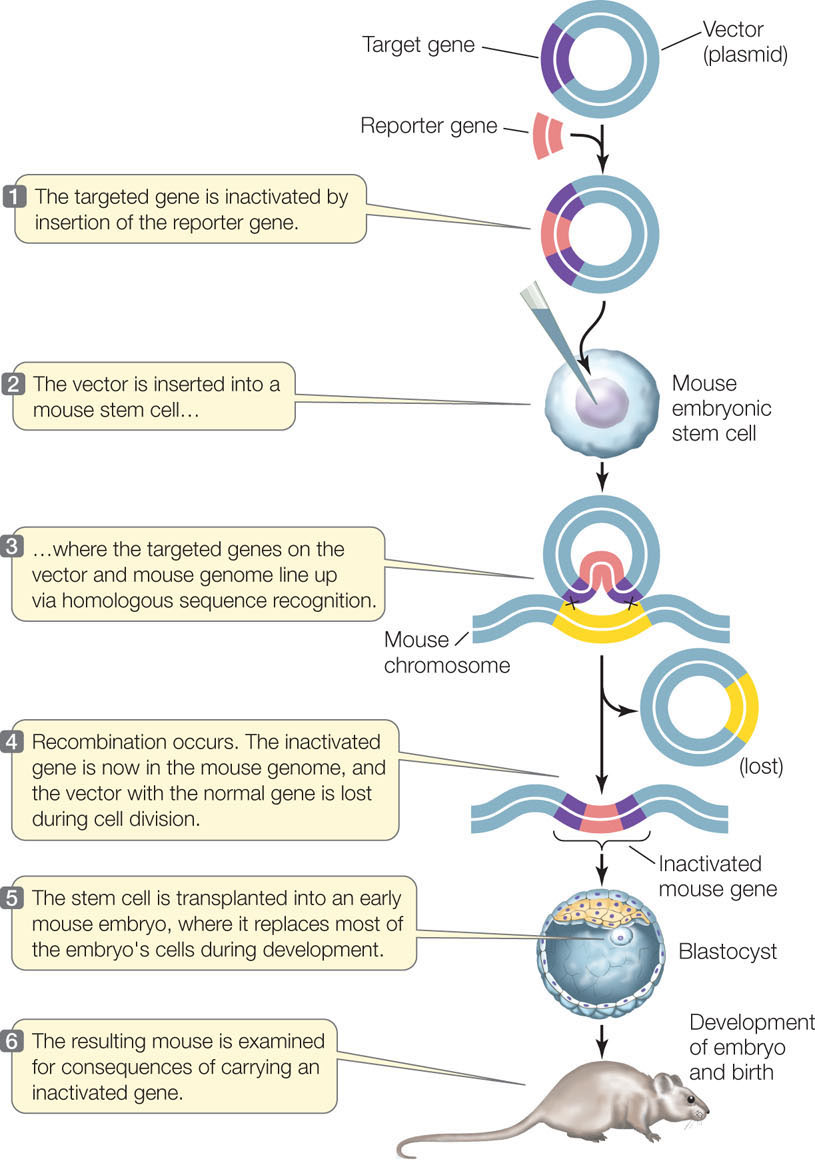
Once the recombinant plasmid has been made, it is used to transfect mouse embryonic stem cells. A stem cell is an unspecialized cell that divides and differentiates into specialized cells. The gene sequences in the plasmid tend to line up with their homologous sequences in the mouse chromosome. If recombination occurs, the disrupted, inactive allele is “swapped” with the functional allele in the cell.
The knockout technique has been important in assessing the roles of many genes, and is especially valuable in studying human genetic diseases. Many such diseases (including phenylketonuria; see Concept 10.1) have knockout mouse models: mouse strains with similar diseases that were produced by homologous recombination. These models can be used to study the diseases and to test potential treatments.
Complementary RNA can prevent the expression of specific genes
Another way to study the expression of a specific gene is to block the translation of its mRNA. This is yet another example of scientists imitating nature. As described in Concept 11.4, gene expression can be controlled in nature by the production of short, single-stranded RNA molecules (microRNAs or miRNAs) that inhibit the translation of target mRNA sequences. Many complex eukaryotes also produce small interfering RNAs (siRNAs), which are short (20–25 bp) double-stranded RNAs derived from much longer double-stranded RNA molecules. As in the production of miRNAs, these double-stranded siRNA molecules are processed into single-stranded molecules, and then each one is guided by a protein complex to a complementary region on an mRNA. The protein complex then catalyzes the breakdown of the targeted mRNA (FIGURE 13.9). These mechanisms for preventing mRNA translation are called RNA interference (RNAi).
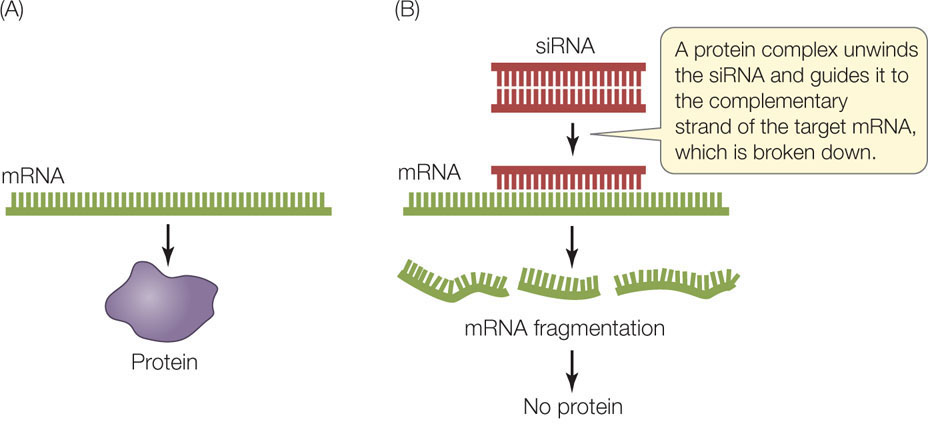
MicroRNAs and siRNAs are examples of antisense RNA because they bind by base pairing to the “sense” bases on the target mRNAs. siRNAs target specific mRNA molecules (from specific genes) because their sequences exactly match the target sequences in the mRNAs. By contrast, miRNAs do not match their targets perfectly, and therefore each one can reduce the expression of multiple, partially matching genes.
RNAi was discovered in the late 1990s, and since then scientists have used synthetic, single-stranded antisense RNAs and double-stranded siRNAs to inhibit the expression of known genes. This technique has been used extensively to block expression of specific genes in the laboratory, as well as in applied situations. For example, macular degeneration is an eye disease that results in near blindness when blood vessels proliferate in the eye. The signaling molecule that stimulates vessel proliferation is a growth factor. An RNAi-based therapy is being developed to target this growth factor’s mRNA, and the therapy shows promise for stopping and even reversing the progress of the disease.
264
DNA microarrays reveal RNA expression patterns
The science of genomics faces two major quantitative realities. First, there are very large numbers of genes in eukaryotic genomes. Second, the pattern of gene expression in different tissues at different times is quite distinctive. For example, the cells of a skin cancer at its early stage may have a different set of mRNAs from those of normal skin cells and cells from a more advanced skin cancer.
To find such patterns, scientists could isolate mRNA from a cell and test for the presence of transcripts from each gene by hybridization or RT-PCR. But that would involve many steps and take a long time. It is far simpler to measure expression of every gene in one step. This is possible with DNA microarray technology, which provides large arrays of sequences for hybridization experiments.
A DNA microarray (“gene chip”) contains a series of DNA sequences attached to a solid surface. The array is divided into a grid of microscopic spots, each containing thousands of copies of a particular oligonucleotide. A computer controls the addition of these oligonucleotide sequences in a predetermined pattern. Each oligonucleotide can hybridize with only one DNA or RNA sequence, and thus is a unique identifier of a gene. Many thousands of different oligonucleotides can be placed in a single microarray.
Microarrays can be used to examine patterns of gene expression in different tissues and under different conditions, and they can be used to identify individual organisms with particular mutations. You can visualize the concept of microarray analysis by following the example illustrated in FIGURE 13.10. Most women with breast cancer are treated with surgery to remove the tumor, and then treated with radiation soon afterward to kill cancer cells that the surgery may have missed. But a few cancer cells may survive in some patients, and these cells eventually form new tumors in the breast or elsewhere in the body. The challenge for physicians is to identify patients with surviving cancer cells so they can be treated aggressively with tumor-killing chemotherapy.
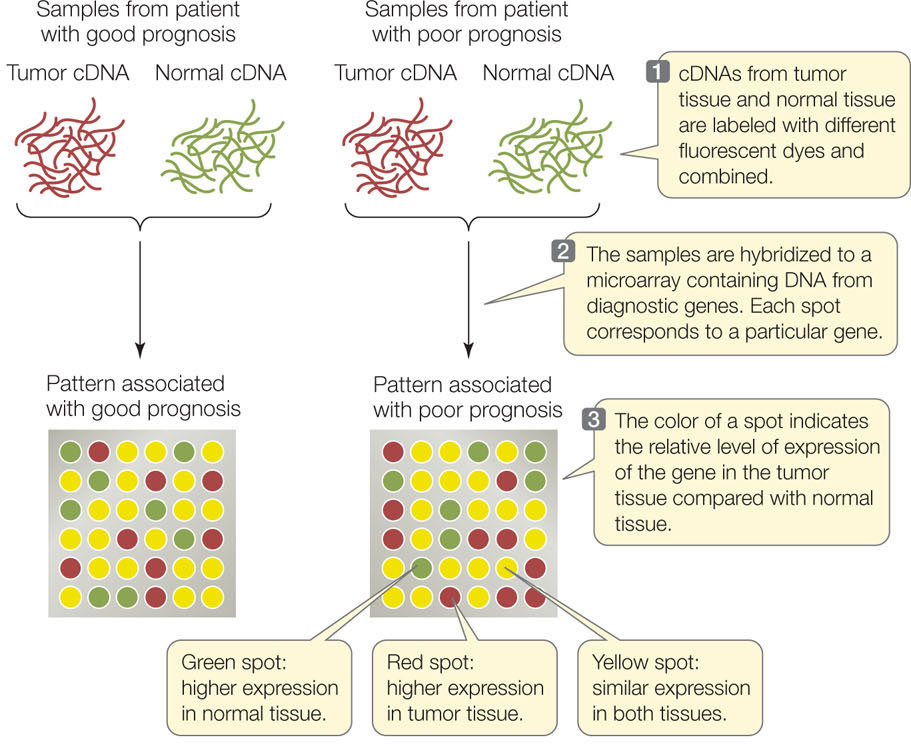
Scientists at the Netherlands Cancer Institute used medical records to identify patients whose cancer recurred or did not recur. They extracted mRNA from the patients’ tumors and made cDNA from the samples. The cDNAs were hybridized to microarrays containing sequences derived from 1,000 human genes. The scientists found 70 genes whose expression differed dramatically between tumors from patients whose cancers recurred and tumors from patients whose cancers did not recur. From this information the Dutch group identified “gene expression signatures” that are useful in clinical decision-making: patients with a good prognosis can avoid unnecessary chemotherapy, whereas those with a poor prognosis can receive more aggressive treatment.
265
CHECKpointCONCEPT13.3
- Outline the steps involved in knocking out a gene in a bacterium and in an animal. What are the uses for these methods?
- What are the differences between a genomic library and a cDNA library?
- You hypothesize that when a corn plant is infected with a fungus, a set of genes is turned on that fights the infection. How would you investigate this hypothesis using microarray technology?
We have now seen how recombinant DNA is made, how cells and organisms are transformed, and how gene expression can be manipulated. In the final concept we will look at some of the many applications of biotechnology.