6.3 RNA STRUCTURE
The discovery that RNA molecules play key roles in converting the genetic information contained in DNA into the proteins that perform structural and catalytic functions in cells motivated the quest to determine the molecular structure of RNAs. In the early 1970s, Alexander Rich, Aaron Klug, and Sung-Hou Kim independently solved the structures of transfer RNAs, revealing how tRNAs carry the amino acids that are used in protein synthesis on the ribosomes. The field of RNA structural biology then languished for almost 20 years, due in part to the technical difficulty of preparing RNA samples for study in the laboratory and the lack of awareness of the wide variety of biological functions of RNA.
The situation changed in 1990, driven by the discoveries of many new kinds of RNA molecules in cells and viruses. In fact, many viral genomes, including those of HIV, HCV, and the influenza viruses, are made entirely of RNA, which can be either single-stranded or double-stranded. Questions about the stability of RNA genomes and how their structures might differ from those of genomic DNA increased the urgency of discovering how RNA molecules are structured.
RNAs Have Helical Secondary Structures
The wide-ranging functions of RNA reflect a structural diversity much richer than that observed in DNA molecules. The propensity of RNA to form compact folded shapes was first revealed in the 1970s with the determination of the molecular structures of several tRNA molecules (Figure 6-22). These analyses showed that a single strand of RNA folds back on itself to form short base-paired or partially base-paired segments connected by unpaired regions (Figure 6-23). This property, called RNA secondary structure, enables RNA molecules to fold into many different shapes that lend themselves to many different biological functions.
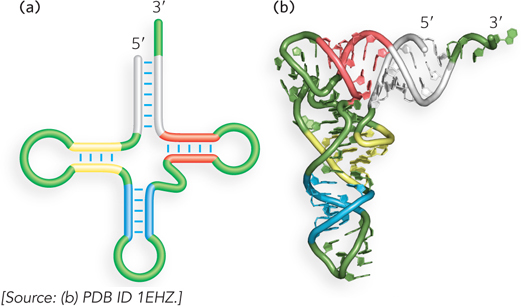
Figure 6-22: The three-dimensional structure of tRNA. (a) The secondary structure of tRNA forms a cloverleaf containing four helices that meet at a central junction. The structure contains several non-Watson-Crick base pairs and loops. (b) The three-dimensional structure of tRNAPhe (a tRNA specific for phenylalanine in protein synthesis), determined by x-ray crystallography, is shown as a ribbon diagram.
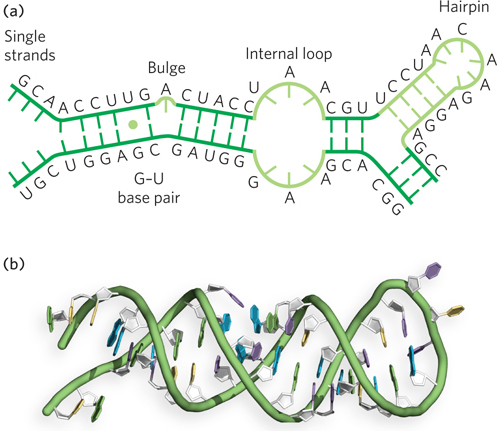
Figure 6-23: Double-helical characteristics of RNA. (a) Some of the diversity in the secondary structure of RNA is shown in these examples of G–U base pairs, bulges, internal loops, and hairpin loop structures. (b) An RNA helix in the form of a hairpin structure similar to the one shown in (a); notice how the unpaired bases at the hairpin loop are incorporated into the structure while maintaining the overall helical geometry of an A-form duplex.
As in DNA, the paired strands in RNA are antiparallel and tend to assume a right-handed helical conformation dominated by base-stacking interactions. Unlike DNA, however, the base-paired segments of RNA are interspersed with a variety of other, non-Watson-Crick, base pairings such as A–A and G–U (Figure 6-24; also see the How We Know section at the end of this chapter). In addition, RNA secondary structures include regions of unpaired nucleotides, which can interact with noncontiguous sequences to stabilize the three-dimensional folding. Such interactions produce compact shapes containing surfaces or crevices that bind other molecules or form sites capable of catalyzing chemical reactions, much like protein enzymes.
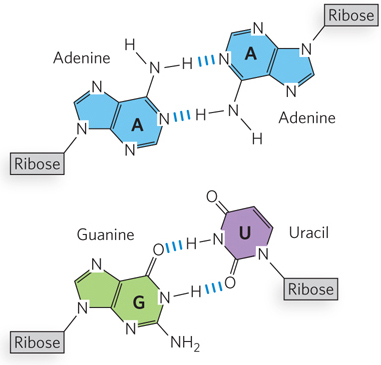
Figure 6-24: A–A and G–U base pairs in RNA.
The greater structural variety in RNA relative to DNA reflects the three main chemical differences between the two polynucleotides: the pentose (2′-deoxyribose in DNA vs. ribose in RNA), the base composition (thymine in DNA vs. uracil in RNA), and the sugar pucker of the pentose (C-2′ endo in DNA vs. C-3′ endo in RNA). The presence of the 2′-hydroxyl group on the sugar of RNA nucleotides provides an extra site for hydrogen bonding, potentially stabilizing three-dimensional folding. This hydroxyl group also influences the sugar pucker, leading to more closely spaced phosphates on the 5′ and 3′ sides of each sugar and hence to a more compact, A-form helical structure (see Figure 6-17).
As in the DNA double helix, RNA base stacking is made energetically favorable by the resulting burial of hydrophobic surfaces away from the hydrophilic surroundings. In tRNA, as well as in the more recently discovered structures of catalytic RNA molecules and ribosomes, virtually all of the bases are stacked, even when they are not part of Watson-Crick base pairings. In each case, structural motifs, such as base-triple interactions (see Figure 6-21a, b) and helix-helix packing, allow stable three-dimensional folding.
Double-stranded RNAs, such as those that form the genomes of some viruses, do exist in nature. These RNAs exist as long helical structures analogous to the DNA double helix. In addition, some RNAs do not seem to form stable three-dimensional structures through local base-pairing interactions. For example, mRNAs seem to perform their function as transient carriers of genetic information without adopting any specific three-dimensional structure. These RNAs may fold into three-dimensional structures only in the presence of bound proteins, forming complexes called ribonucleoproteins (RNPs).
RNAs Form Various Stable Three-Dimensional Structures
Most of the highly structured RNAs contain noncanonical base pairs and backbone conformations not observed in DNA. In many cases, the 2′-hydroxyl group on ribose, a chemical feature that distinguishes RNA from DNA, seems to be directly or indirectly responsible for these unique structural properties. Recall that the presence of this 2′ hydroxyl gives ribose its C-3′ endo geometry (its sugar pucker), as distinct from the C-2′ endo characteristic of deoxyribose. This seemingly small difference in chemical conformation leads to a distinct helical geometry for RNA relative to DNA. Furthermore, it enables direct or water-mediated hydrogen bonding between the 2′-hydroxyl of one RNA nucleotide and the adjacent ribose or phosphate (Figure 6-25). As a result, RNA helices are more thermodynamically stable than DNA helices of the same length and sequence.
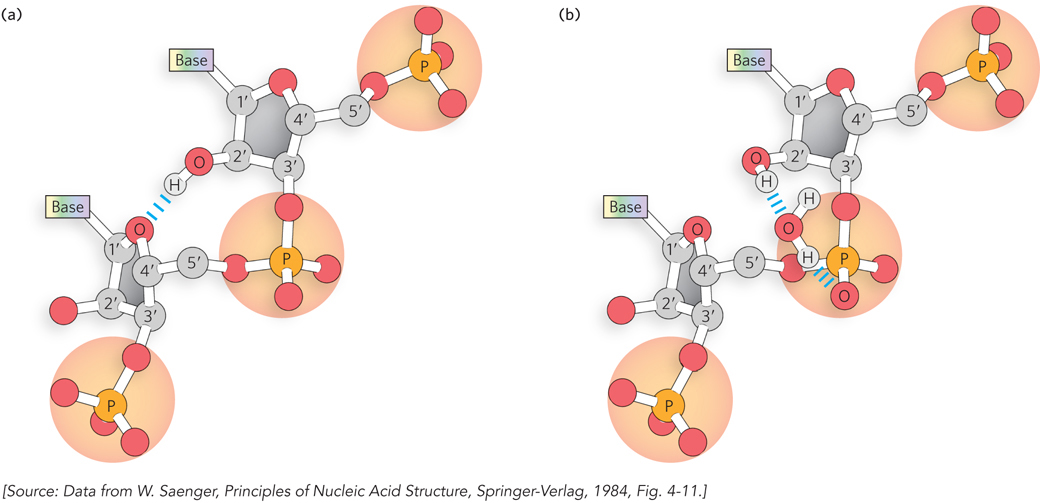
Figure 6-25: Stabilization of RNA secondary structure. The 2′-hydroxyl group on ribose causes RNA to favor the C-3′ endo sugar pucker, due to (a) direct hydrogen bonding between a 2′ hydroxyl on one ribose and the ring oxygen on an adjacent ribose, and (b) hydrogen bonding, through a water molecule, between a 2′ hydroxyl and a phosphate oxygen.
Where complementary sequences are present in an RNA molecule, the predominant double-stranded structure in the RNA is an A-form, right-handed double helix (see Figure 6-17). The A-RNA helix has a wider, shallower minor groove and a narrower, deeper major groove than the B-form helix observed for most DNA. The A-form geometry in RNA has a shorter distance between adjacent phosphates in the sugar–phosphate backbone than in B-DNA, a consequence of the C-3′ endo sugar pucker of ribose. A B-form RNA helix has not been observed in nature. Z-RNA helices have been induced to form in the laboratory under high-salt or high-temperature conditions but are not known to occur in cells.
As mentioned previously, mismatched or unmatched bases are common in base-paired segments of RNA, locally disrupting the regular A-form helix and resulting in bulges or internal loops (see Figure 6-23). The potential for base pairing within a single strand of RNA frequently produces thermodynamically stable secondary structures that consist of hairpinlike conformations capped by connecting loops. Such hairpins are the most common type of RNA secondary structure, often containing specific short sequences at their ends (such as UUCG or GAAA) that form particularly energetically favorable loops. The nucleotides in the loops are arranged to maximize hydrogen bonding and base stacking, thereby enhancing thermodynamic stability. Important additional structural contributions are made by hydrogen bonds that are not part of canonical Watson-Crick base pairs. These properties, evident in the structures of tRNAs and catalytic RNAs, were also evident in the ribosome structures solved by Jamie Cate and others (see this chapter’s Moment of Discovery). The functions of these highly structured RNAs, like those of proteins, depend on their three-dimensional properties.
Weak interactions, especially base-stacking interactions, play a major role in stabilizing RNA structures, just as they do in DNA. The 2′ hydroxyl is often involved in hydrogen bonds and van der Waals interactions that stabilize alternative helical shapes and conformations, such as loops and kinks that require the close approach of the two phosphodiester backbones. Divalent and monovalent metal ions—such as Mg2+, Ca2+, K+, and Na+—bind to specific sites in RNA and help shield the negative charge of the backbone, allowing parts of the molecule to pack more tightly together (Figure 6-26).
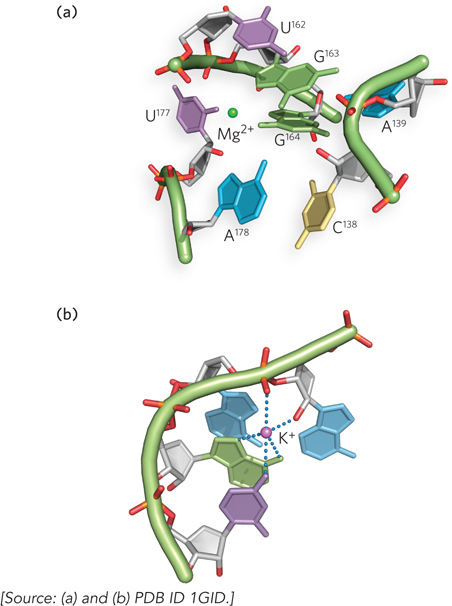
Figure 6-26: Mg 2+ and K+ binding in RNA structure. (a) Divalent magnesium ions (Mg2+) coordinate to phosphate groups and stabilize the close approach of phosphate backbones in the folded structure of the P4–P6 domain of the Tetrahymena group I ribozyme (a catalytic RNA). (b) Monovalent potassium ions (K+) bind to specific sites in the P4–P6 domain, where they favor interactions between both backbone atoms and bases.
The diversity of RNA three-dimensional structures found in nature is illustrated by riboswitches, non-protein-encoding RNA sequences with the unique ability to directly bind cellular metabolites and form alternative metabolite-free and metabolite-bound shapes. Found widely among bacteria and sporadically in other organisms, riboswitches can change their structure on binding to a particular metabolite and, as consequence, alter the efficiency of synthesis of proteins involved in pathways utilizing that metabolite. Such metabolite-sensing RNAs function through complex three-dimensional folding to create structures that can selectively recognize a small molecule within the mixture of molecules in a cell. Due to their ability to adopt a wide range of structures, riboswitches can bind to molecules ranging in size and type from tiny ions to large organic molecules. Although riboswitches employ common structural principles to form their metabolite-binding shapes, different riboswitch classes possess unique features for ligand recognition, even those that have evolved to recognize the same metabolites (Figure 6-27). We will encounter further examples of the wide-ranging structures of RNA when discussing ribosomes and catalytic introns in Chapters 16 and 18.
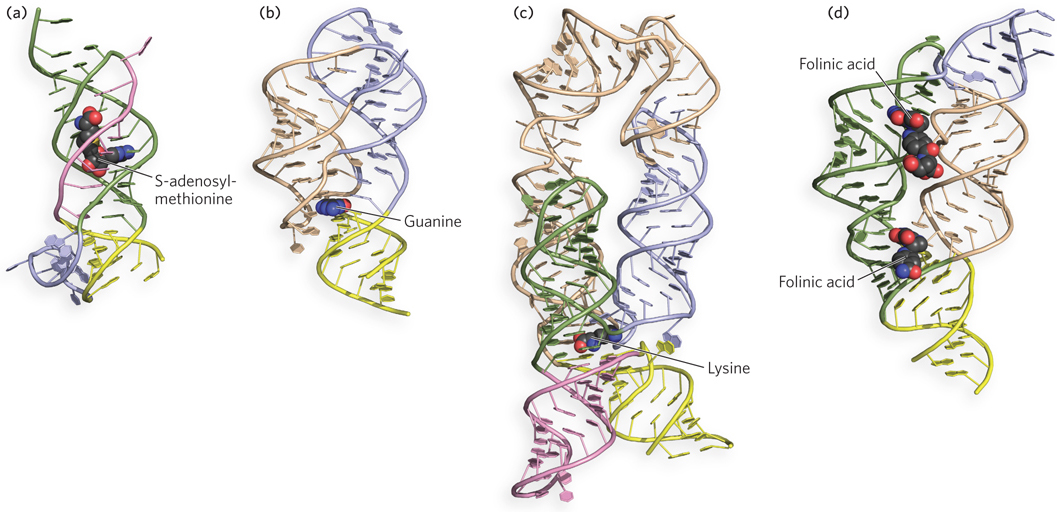
Figure 6-27: Examples of the diversity of RNA three-dimensional structures in riboswitches. The phosphodiester backbone in each case is shown as a solid tube; bound metabolites are shown in red. (a) The S-adenosylmethionine-II riboswitch; (b) a guanine-binding riboswitch; (c) a lysine-binding riboswitch; and (d) a tetrahydrofolate-binding riboswitch, bound here to two molecules of folinic acid.
The analysis of RNA structure, along with the relationship between structure and function, is an emerging field of inquiry with many of the same complexities as the analysis of protein structure. The importance of understanding RNA structure has grown as we have become increasingly aware of the large number of functional roles for RNA molecules. For example, we now know that the extensive structural features found in the genomic RNA of HIV control viral gene expression (Highlight 6-2).
SECTION 6.3 SUMMARY
RNA is chemically better suited than DNA to forming stable three-dimensional folds, due to the 2′-hydroxyl group of ribose in the RNA backbone. The presence of the 2′ hydroxyl makes RNA vulnerable to hydrolysis, but it also allows additional hydrogen bonding between segments of the molecule.
RNA molecules can exist as long double-helical structures, typical of viral genomic RNA, or, more commonly, as single strands that fold up into short helical regions connected by loops and unpaired segments.
Base-paired segments of RNA generally adopt the compact geometry of A-form helices. This structure arises because RNA favors a different sugar pucker from that found in DNA (C-3′ endo in RNA vs. C-2′ endo in DNA), which causes the phosphates in the backbone to become more closely spaced than in B-DNA. In some large RNAs, short helices undergo RNA-RNA and RNA–metal ion interactions to form complex three-dimensional structures.
Base pairs other than canonical A=U and G≡C pairs are common in RNAs, including A–A and G–U. In all cases, base pairs or single bases are most stable when stacked on top of one another in a helix.
Divalent and monovalent metal ions (Mg2+, Ca2+, K+, and Na+) bind to specific sites in RNA and help shield the negative charge of the backbone, allowing parts of the molecule to pack more tightly together.
HIGHLIGHT 6-2 MEDICINE: RNA Structure Governing HIV Gene Expression

Kevin Weeks
RNA structures can be investigated by testing for reactivity to various chemical reagents. This is possible because bases that are paired or are folded inside the RNA, or those that are conformationally rigid, are protected against modification or cleavage by reactive chemicals. By analyzing which sites in a folded RNA molecule are resistant or sensitive to chemical reagents, researchers can obtain information about its structural features.
Kevin Weeks and his colleagues at the University of North Carolina used this approach to analyze the role of RNA structure in regulating gene expression in HIV-1 (a strain of the human immunodeficiency virus). Viral RNA was extracted from viral particles (virions) under gentle conditions, such that the native structure was preserved. The RNA was then treated with a chemical reagent, 1-methyl-7-nitroisatoic anhydride (1M7), which preferentially acylates conformationally flexible nucleotides at the 2′-OH of ribose. After the RNA was allowed to react with 1M7, acylated sites were detected by reverse transcription. This procedure uses the purified enzyme reverse transcriptase to make a DNA copy of the viral RNA. The researchers found that acylation blocked the progression of polymerization, generating a truncated DNA fragment extending up to the site of acylation in the RNA sequence. Following fractionation of the resulting DNAs, the sites of acylation were detected and mapped onto the HIV-1 genome sequence.
Analysis of these data revealed that in addition to encoding viral proteins in its nucleotide sequence, the viral genomic RNA is also three-dimensionally structured to optimize protein production (Figure 1). Highly structured regions in the RNA—and hence a dearth of experimentally acylated sites—occur within sequences that encode interprotein linkers, or loops, between protein domains. This finding implies that RNA structure slows down the rate at which ribosomes move through these parts of the protein-coding sequence, providing time for the newly synthesized viral proteins to fold into their active structures. The results of this study underscore the idea that the HIV genome and, perhaps, the mRNAs contain structured regions that regulate expression of the proteins they encode. RNA secondary and higher-order structure may therefore constitute an important component of the genetic code.
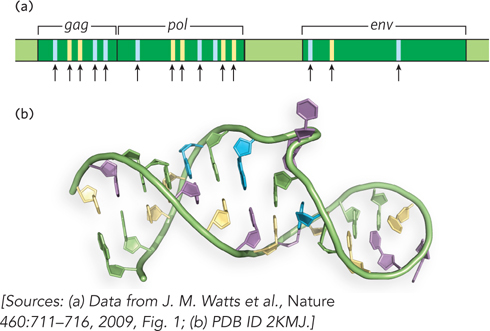
FIGURE 1 The organization and structure of the HIV-1 genomic RNA influences viral protein expression. (a) This portion of the HIV-1 genome shows three open reading frames, gag, pol, and env, that are translated into long polyprotein precursors; proteolytic cleavage generates multiple proteins from each reading frame. Blue bars represent sites that code for interprotein linkers, and yellow bars indicate sites coding for loops between protein domains. Black arrows mark regions of the RNA genome that are highly structured; note that these areas coincide with areas that link proteins or protein domains. (b) An example of RNA secondary structure in the HIV-1 genome.







