13.2 ENZYMATIC MACHINES IN BACTERIAL RECOMBINATIONAL DNA REPAIR
The complex pathways shown in Figures 13-2 through 13-6 have been elucidated by genetic studies carried out in bacteria and eukaryotes. Many of the enzymes that promote the steps in these pathways have been characterized in vitro, and multiple steps of some pathways have been reconstituted with purified enzymes (Table 13-1). Complete reconstitution of a DSB repair or a replication fork repair remains a key goal of the field. In this section we describe the major enzyme systems found in bacteria, before moving on to eukaryotic recombination systems.
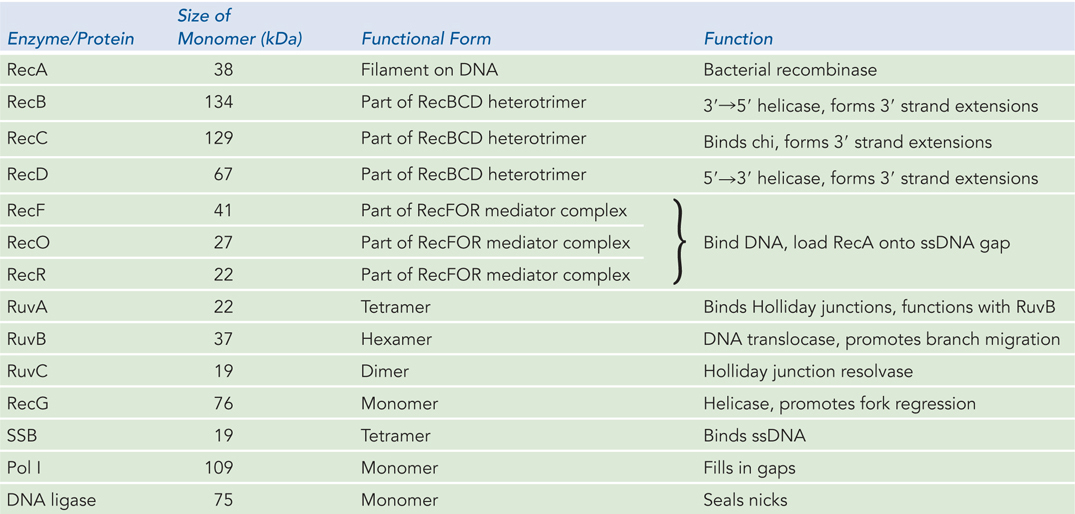
RecBCD and RecFOR Initiate Recombinational Repair
In all cells, the process of recombinational DNA repair revolves around a recombinase enzyme. In bacteria, that recombinase is the RecA protein. Before RecA can act, the stage must be set by other enzymes that degrade one strand of DNA, where necessary, and load RecA onto the DNA. The RecBCD and RecFOR complexes, with three subunits each, are the initiators of recombinational DNA repair. RecBCD loads the RecA protein onto DNA to repair double-
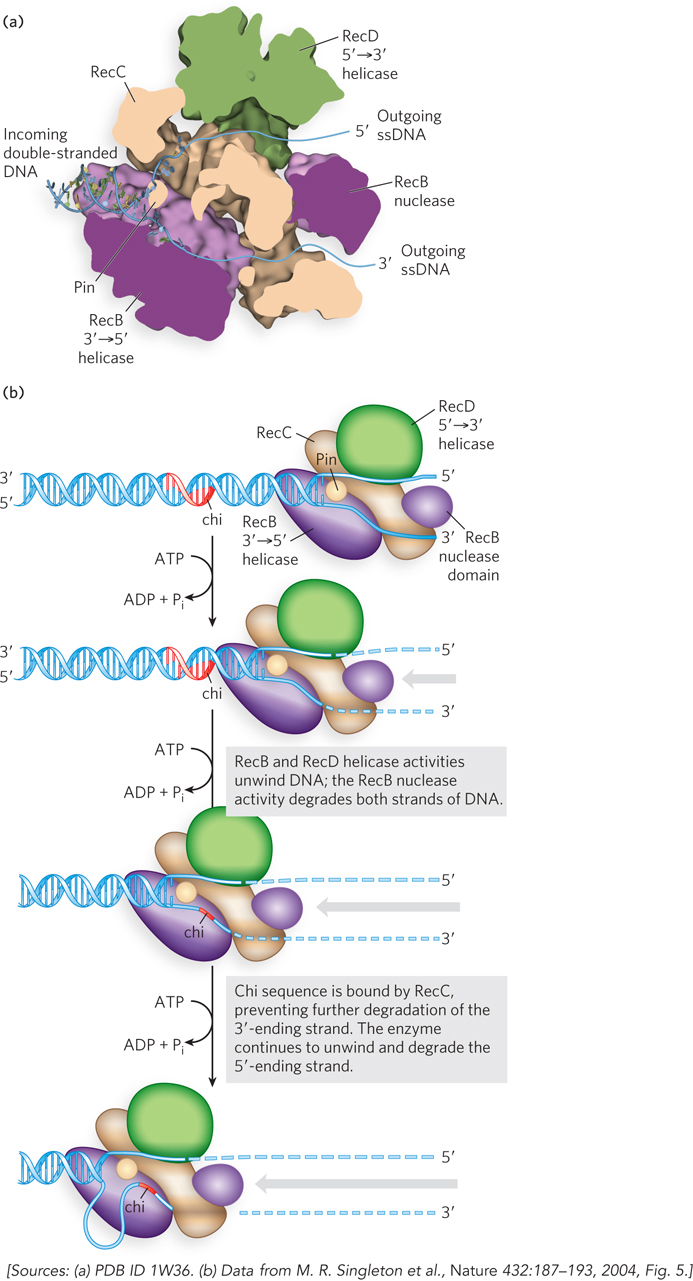
In E. coli, the recB, recC, and recD genes encode the heterotrimeric RecBCD enzyme, which has both helicase and nuclease activities. This enzyme has two jobs: (1) it prepares the 3′-ending single strand by degrading the 5′-ending strand at the site of a DSB, and (2) it directly loads the RecA protein onto the prepared single-
458
As the 3′-ending strand is turned into a single strand, it is rapidly bound by the single-
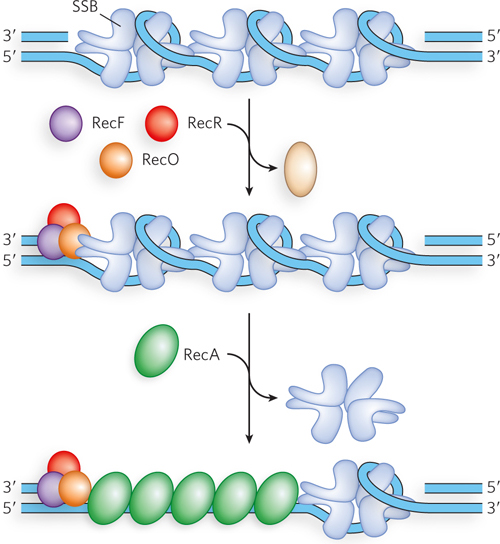
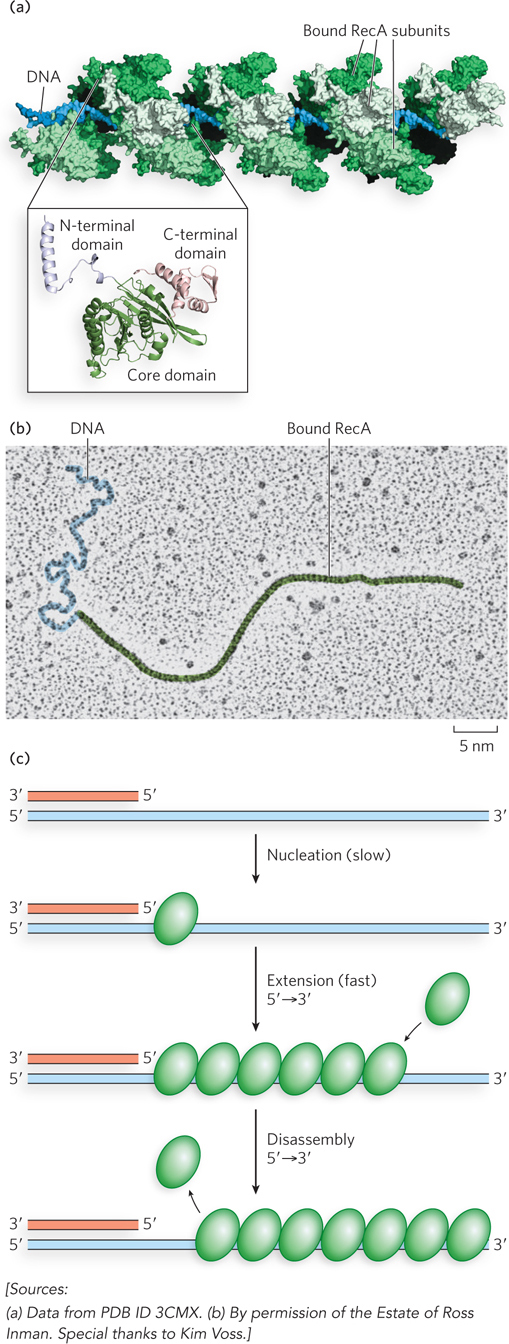
As we will see, the RecA protein forms a nucleoprotein filament on the DNA. The filament is formed in two steps: first the binding of a few RecA subunits (called nucleation), then extension of the filament from that initial complex. The nucleation step limits the overall process and is the step that is blocked by SSB. As the RecBCD enzyme unwinds the DNA and degrades the 5′-ending strand, a domain within the RecB subunit recruits RecA to the 3′-ending strand. This loading function of RecB serves to nucleate the RecA binding that is needed to promote strand invasion.
The RecA protein must also be loaded onto single-
459
RecA Protein Is the Bacterial Recombinase
The bacterial RecA protein is the prototype of a class of proteins found in all organisms, from bacteria to humans (see the How We Know section at the end of this chapter). The RecA-
RecA-
460
The RecA filament can facilitate strand exchange with a variety of substrates in vitro. The single-
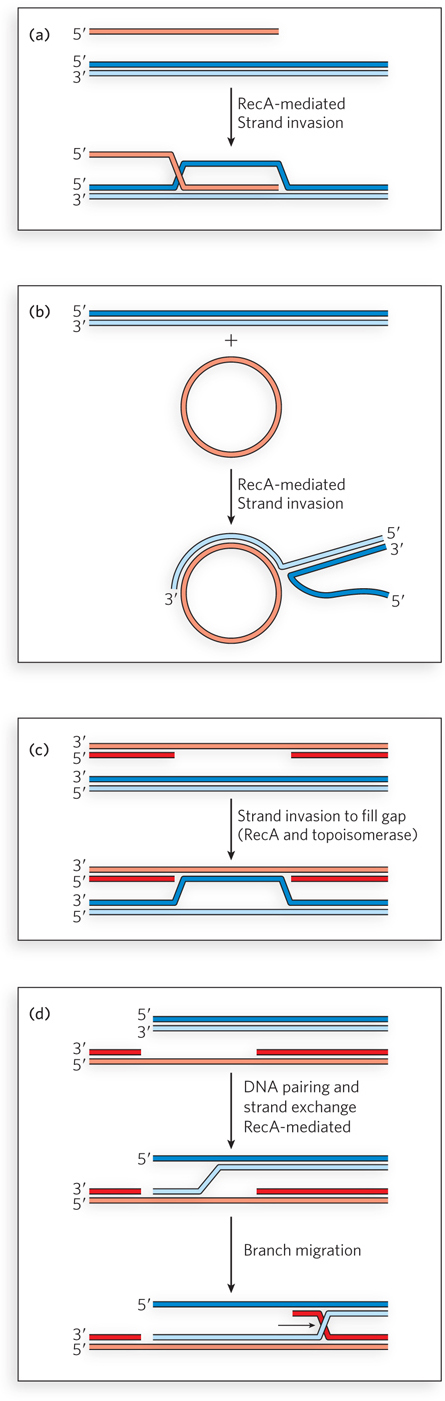
In each scenario, the single strand of DNA is first bound by RecA to establish the nucleoprotein filament. The RecA filament then promotes strand invasion into a homologous double-
When purified RecA protein promotes DNA strand exchange in vitro between a circular single strand and a linear duplex, the substrates and products have distinctive structures that are readily separated and visualized by agarose gel electrophoresis (Figure 13-11). The initial pairing of the RecA-
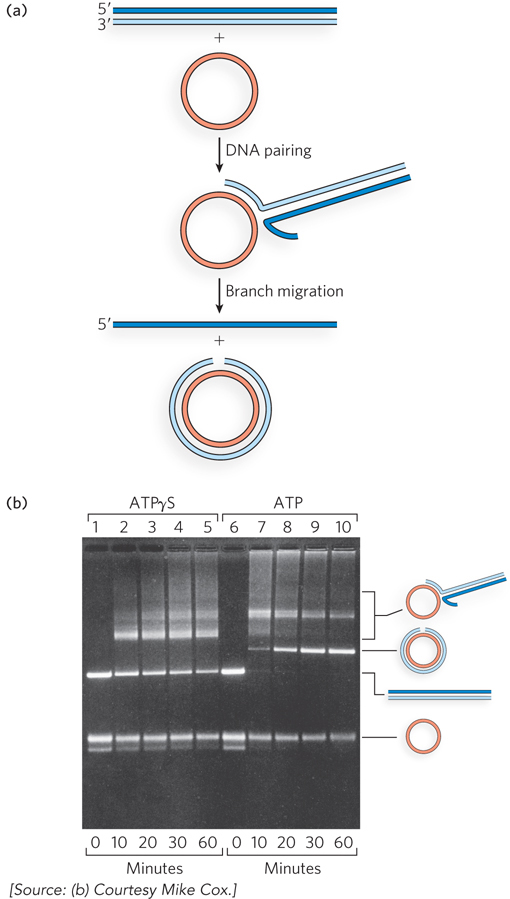
461
If the circular substrate is a gapped double-
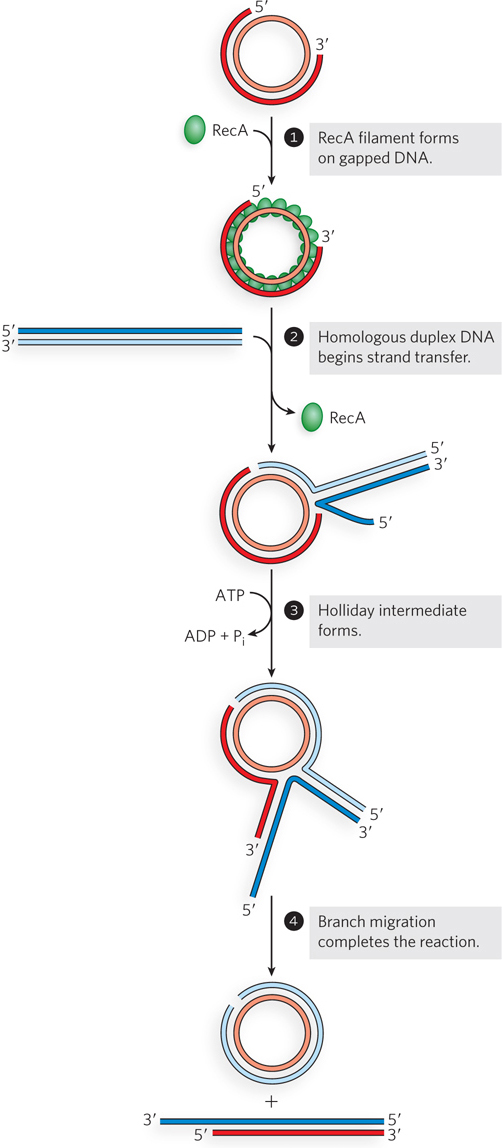
RecA Protein Is Subject to Regulation
In principle, RecA-
Regulation occurs at three levels: transcription of the recA gene, autoregulation, and regulation by other proteins. Transcriptional regulation occurs within the context of the bacterial SOS response (described in Chapter 20). Most regulation at other levels is directed at the formation, disassembly, and function of RecA protein filaments.
462
Autoregulation is “self” regulation. The RecA protein suppresses its own activities by means of a highly charged C-
463
Many other proteins play a role in regulating the RecA protein. As we have seen, the RecBCD and RecFOR complexes facilitate the RecA filament nucleation process. Reliance on these loading functions helps direct RecA filament formation to DNA regions where it is needed. Another regulatory protein, RecX, binds to the growing RecA filament end and halts filament extension. A protein called DinI binds along the RecA filament and stabilizes it, while at the same time limiting the DNA strand exchange process. The helicase UvrD actively removes RecA filaments from the DNA when they are no longer needed. These and other proteins, working as an integrated system, help limit RecA function and direct it toward particular repair requirements.
Multiple Enzymes Process DNA Intermediates Created by RecA
RecA is not the only protein in a bacterial cell that can promote branch migration; other enzyme systems are specialized for that task. As one example, the processing of Holliday intermediates is facilitated by a complex called RuvAB (repair of UV damage). Up to two RuvA protein tetramers bind to a Holliday intermediate and form a complex with two RuvB hexamers (Figure 13-13). The donut-
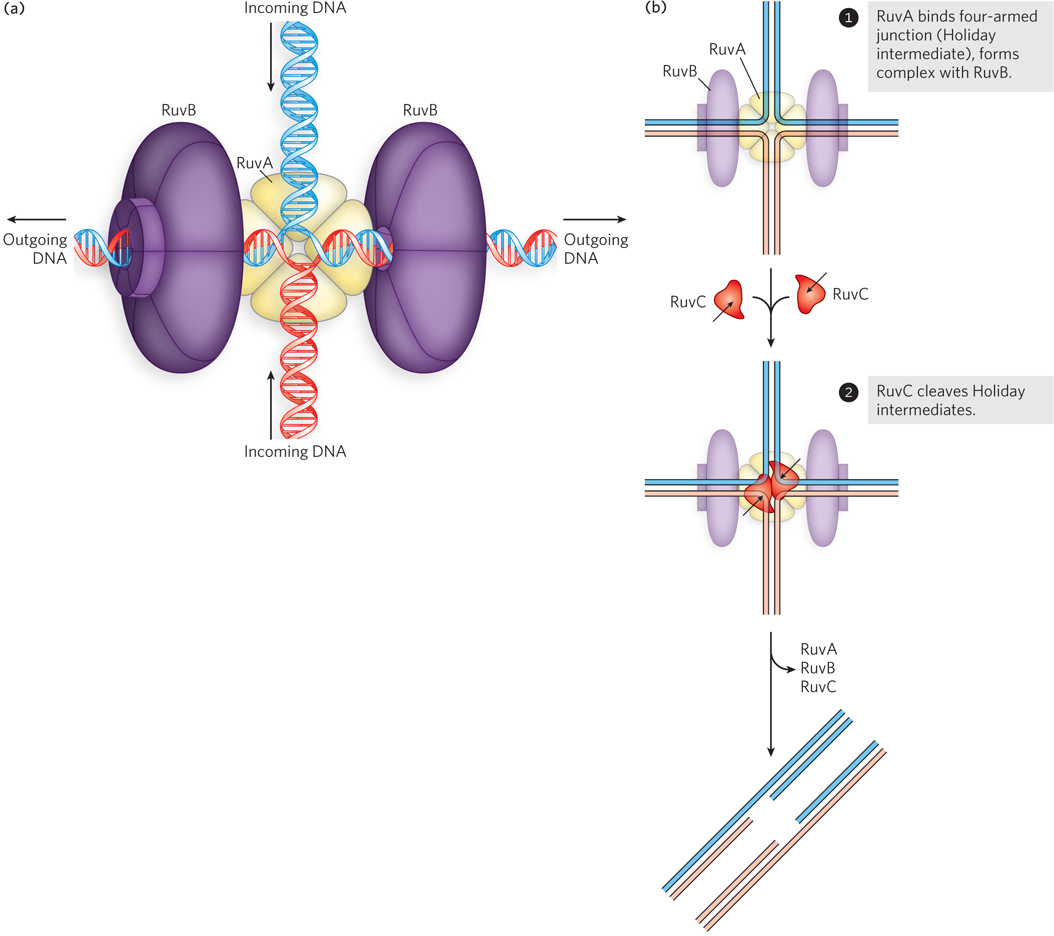
464
HIGHLIGHT 13-1 EVOLUTION:A Tough Organism in a Tough Environment: Deinococcus radiodurans
Some radioactive isotopes, such as 60Co and 137Cs, emit a type of ionizing radiation called γ rays. Gamma rays are photons; they transmit energy to atoms in solution, generating ions that include the highly reactive hydroxyl radical. In a living cell exposed to γ rays, any molecule can be damaged, including proteins and DNA. Double-
The energy deposited by electromagnetic radiation is measured in rads or grays (1 Gy = 100 rads). For a human cell, a dose of 2 to 5 Gy is lethal. In the 1950s, it became clear that some organisms are surprisingly resistant to radiation. For example, in efforts to use radioactive sources to sterilize food, some sealed food samples were spoiled by bacterial action even after exposure to γ radiation at levels up to 4,000 Gy. The culprit was a pink, non-
The Deinococcus genome consists of four circular DNA molecules, all generally present in multiple copies. After γ irradiation, the cells stop growing and DNA repair begins. Overlapping DNA fragments are spliced together, and the entire genome is accurately reconstituted within a few hours. The cells begin to grow and divide again as if nothing had happened. It is perhaps the most remarkable feat of DNA repair we know of so far.
This process is demonstrated in the gel shown in Figure 1. Following various treatments, D. radiodurans genomic DNA was isolated, treated with a restriction enzyme that cuts only a few times in the genome, and subjected to pulsed field gel electrophoresis (see Chapter 7). In cells grown under normal conditions, this procedure yields the series of large DNA fragments shown in the second lane of the gel (the first lane consists of markers of known molecular weight). Immediately after γ irradiation at 5,000 Gy, this banding pattern disappears, replaced by a smear of randomly sized, smaller DNA fragments. Over the next 3 to 4 hours, the normal band pattern reappears as the genome is accurately reconstituted.
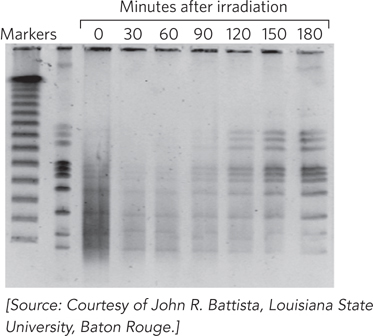
Genome reconstitution in D. radiodurans is recombinational DNA repair on a massive scale. The Deinococcus RecA protein (DrRecA) plays a key role, and most of the radiation resistance of this organism disappears if DrRecA is inactivated. There are two stages of repair, each requiring about 90 to 120 minutes under optimal conditions. The first stage uses a process similar to synthesis-
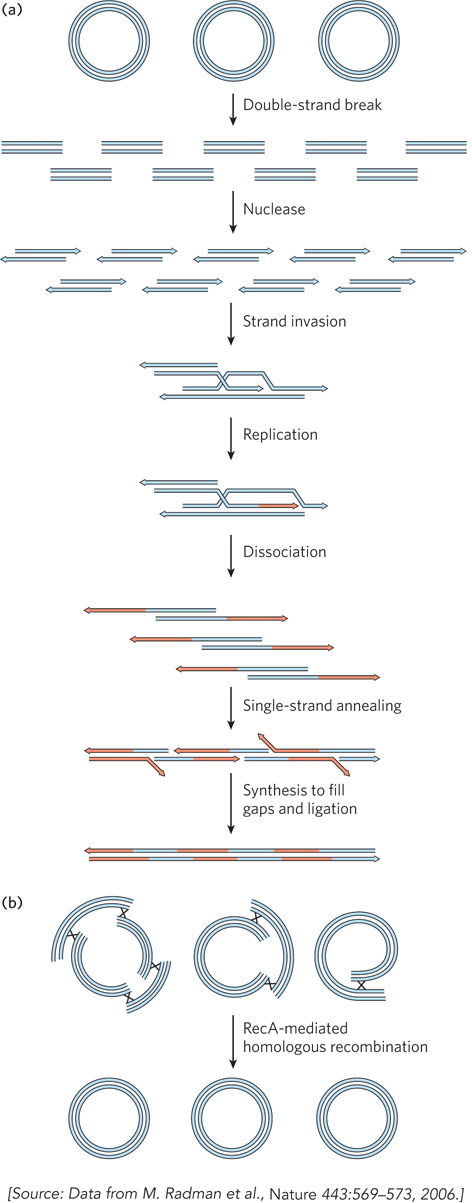
For several decades, D. radiodurans was considered the most radiation-
465
The bacterial recombination systems that repair replication forks can also repair DSBs created by ionizing radiation, sometimes with startling proficiency. The bacterium Deinococcus radiodurans can survive and prosper after absorbing doses of ionizing radiation sufficient to generate thousands of DSBs (Highlight 13-1).
The repair of stalled or collapsed replication forks is generally followed by a restart of replication. A five-
466
Repair of the Replication Fork in Bacteria Can Lead to Dimeric Chromosomes
Some pathways of replication fork repair lead to the creation of a Holliday intermediate behind the reconstituted fork instead of in front of it (Figure 13-14; see also Figure 13-5). This Holliday intermediate can be resolved by RuvC in one of two ways: by cleaving the crossover strands (shown as resolution path X in Figure 13-14) or by cleaving the template strands (resolution path Y). The first path, cleaving the crossover strands, simply leads to the completion of replication and the segregation of two monomeric chromosomes into daughter cells. The second, cleaving the template strands, has a special consequence when the genome is circular, as it is for most bacterial chromosomes: it ultimately creates a single dimeric chromosome that cannot be segregated at cell division. Under normal growth conditions, this outcome is observed in about 15% of cells in an E. coli culture.
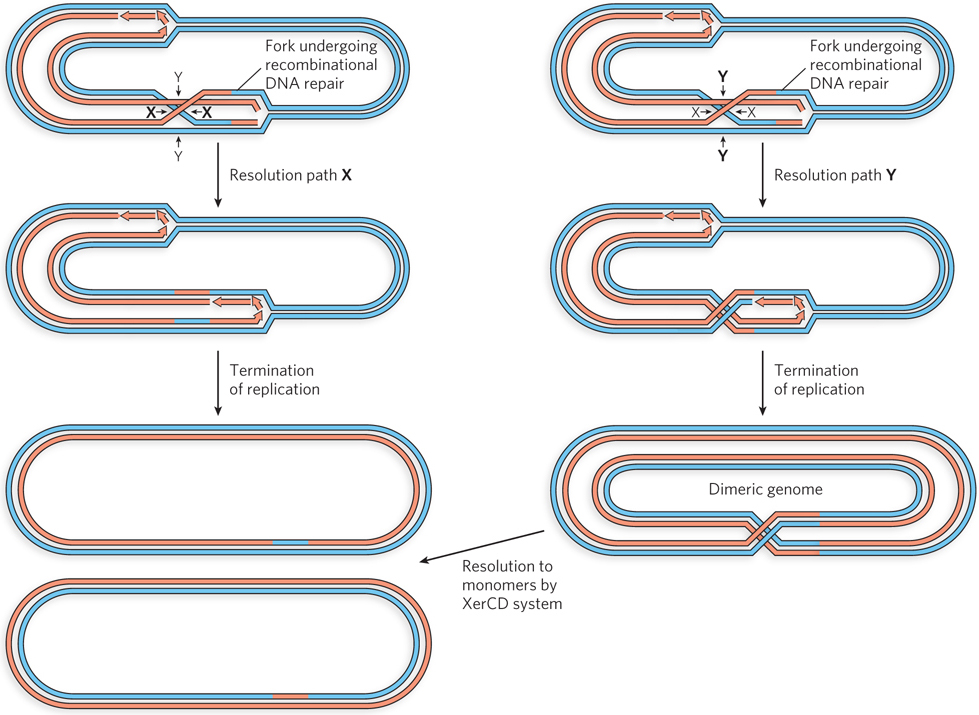
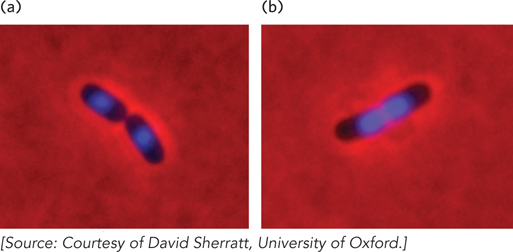
Cells harboring dimeric chromosomes do not die. Instead, the stalled chromosomal segregation is detected, triggering the activity of a specialized site-
SECTION 13.2 SUMMARY
The bacterial RecBCD and RecFOR complexes provide pathways for loading the RecA protein onto single-
stranded extensions (at double- strand breaks) or onto single- strand gaps, respectively. The bacterial RecA protein is the prototypical recombinase, forming a filament on single-
stranded DNA and promoting strand invasion reactions. Recombination is a highly regulated process, and the RecA protein is the major target for regulation, which occurs through transcriptional regulation, autoregulation, and regulation by other proteins.
467
Recombination intermediates generated by RecA are processed by enzymes such as the RuvA, RuvB, and RuvC proteins.
In the circular bacterial chromosome, resolution of Holliday intermediates associated with replication fork repair can lead to the formation of dimeric chromosomes.