
Just as medieval fortresses, such as the Dover Castle, used walls and fortifications to defend their territory, the immune system constantly battles against foreign invaders such as viruses, bacteria, and parasites to defend the organism. Antibody molecules provide a key element in the immune system’s defensive arsenal. For example, specific antibodies can bind to molecules on the surfaces of viruses and prevent the viruses from infecting cells. Above right, an antibody binds to one subunit on hemagglutinin from the surface of influenza virus.
We are constantly exposed to an incredible diversity of bacteria, viruses, and parasites, many of which would flourish in our cells or extracellular fluids were it not for our immune system. How does the immune system protect us? The human body has two lines of defense: an innate immune system that responds rapidly to features present in many pathogens, and an adaptive immune system that responds to specific features present only in a given pathogen. Both the innate and the adaptive immune systems first identify features on disease-causing organisms and then work to eliminate or neutralize those organisms. While a thorough description of the immune system is certainly beyond the scope of this book, this chapter will focus on how biochemical concepts such as protein structure, receptor-ligand interactions, and signal transduction are applied to the identification of pathogens.
The immune system must meet two tremendous challenges in the identification of pathogens: (1) to produce a system of receptors diverse enough to recognize a wide array of potential pathogens and (2) to distinguish invaders and their disease-causing products from the organism’s own products (i.e., self- versus nonself-recognition). To meet these challenges, the innate immune system evolved the ability to recognize structural elements, such as specific glycolipids or forms of nucleic acid, that are well conserved in pathogens but absent in the host organism. The repertoire of such elements is limited, however, and so some pathogens have strategies to escape detection. The adaptive immune system has the remarkable ability to produce more than 108 distinct proteins, called antibodies, that can recognize different foreign molecules, and more than 1012 receptors on immune cells, called T-cell receptors (TCRs), each of which presents a different surface with the potential to specifically bind a structure from a foreign organism. In producing this vast range of defensive molecules, however, the adaptive immune system has the potential to create antibodies and T-cells that recognize and attack cells or molecules normally present in our bodies—a situation that can result in autoimmune diseases.
This chapter will examine these challenges, focusing first on the structures of proteins that recognize foreign organisms and then on the mechanisms for protecting us from a specific pathogen once it has been recognized. The chapter will closely examine the modular construction of the proteins of the immune system—identifying structural motifs and considering how spectacular diversity can arise from modular construction.
Innate immunity is an evolutionarily ancient defense system
Innate immunity is an evolutionarily ancient defense system found, at least in some form, in all multicellular plants and animals. The innate immune system represents the first line of defense against foreign pathogens, relying on common features of invading organisms to identify and eliminate these threats. Components of the innate immune system include the epithelial lining that surrounds host cells and the specialized cells, called phagocytes, that can ingest and destroy pathogens without the aid of the adaptive immune system.
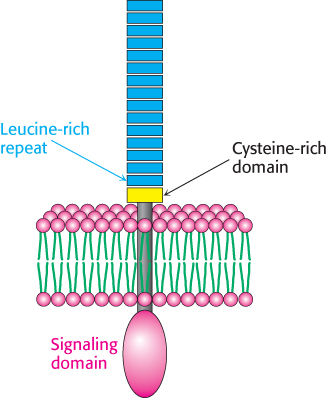
FIGURE 34.1Toll-like receptor. Each receptor is made up of a set of 18 or more leucine-rich repeat sequences, followed by a cysteine-rich domain, a single transmembrane helix, and an intracellular domain that functions in signal transduction.
The innate immune system also includes a family of receptors that can recognize specific features present in most pathogens and yet not respond to materials normally present in the host. The best-understood of these receptors are the Toll-like receptors (TLRs). The name “toll-like” is derived from a receptor known as Toll encoded in the Drosophila genome; Toll was first identified in a screen for genes important for Drosophila development and was subsequently shown to play a key role in the innate immune system. The TLRs have a common structure (Figure 34.1). Each receptor consists of a large extracellular domain built primarily from repeated amino acid sequences termed leucine-rich repeats (LRRs). Each LRR typically contains 20–30 residues, including 6 that are usually leucine. The human TLRs have from 18 to 27 LRR repeats that are capped by a cysteine-rich domain and followed by a sequence forming a single transmembrane helix and an intracellular signaling domain. This signaling domain is not a protein kinase but acts as a docking site for other proteins. Most TLRs are expressed in the cell membrane for the detection of extracellular pathogens such as fungi and bacteria. Other TLRs are located in the membranes of internal compartments for the detection of intracellular pathogens such as viruses and some bacteria.
Each TLR targets a specific molecular characteristic, often called a pathogen-associated molecular pattern (PAMP), found primarily on invading organisms (Table 34.1). Typically, a PAMP is a critical component of the pathogen’s function: mutations in these targets cannot easily block recognition by the TLR without compromising the activity of the pathogen. One particularly important PAMP is lipopolysaccharide (LPS), also referred to as endotoxin, a specific class of glycolipid found in the cell walls of Gram-negative bacteria such as E. coli. LPS is recognized by TLR4. The response of the innate immune system to LPS can be easily demonstrated. Injection of less than 1 mg of LPS into a human being produces a fever and other signs of inflammation even though no living organisms are introduced.
TABLE 34.1 Pathogen-associated molecular patterns (PAMPs) recognized by human TLRs
|
|
|
|
|
|
|
|
|
|
|
Bacteria, viruses, parasites |
|
|
Double-stranded RNA (dsRNA) |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Single-stranded RNA (ssRNA) |
|
|
|
Single-stranded RNA (ssRNA) |
|
|
|
|
Viruses, bacteria, protozoa |
|
|
|
|
Data from: O. Takeuchi and S. Akira, Cell 140:805–820, 2010. |
How do TLRs recognize PAMPs? The extracellular domain from human TLR3 has a remarkable structure that is representative of many other TLRs (Figure 34.2). Each LRR contributes a single β strand to a large parallel β sheet that lines the inside of a concave, hooklike structure. In most cases, the PAMP is recognized by the surface formed between two receptors. The PAMP for TLR3, double-stranded RNA, fits in the groove between two monomers, colored blue and yellow in Figure 34.3. Ligand-induced dimerization of the receptor enables the initiation of a signaling cascade within the cell. We encountered similar examples of signal transduction across a membrane through receptor dimerization in Chapter 14.
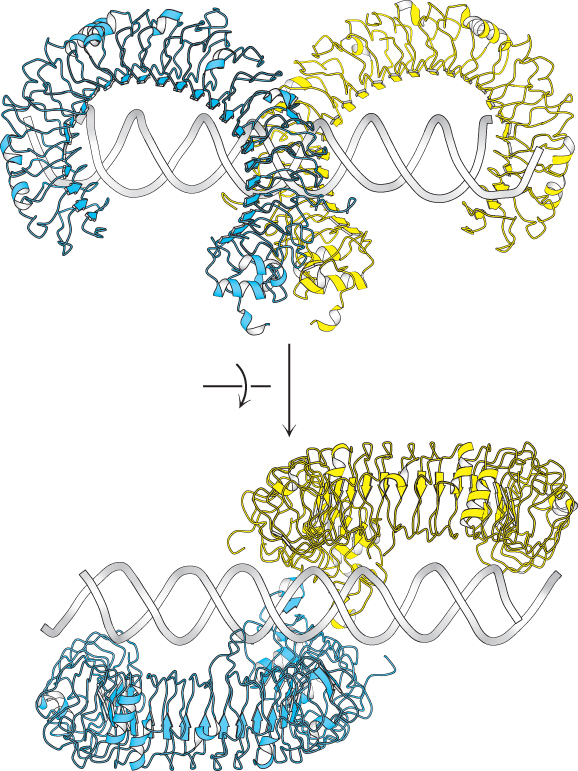
FIGURE 34.3Recognition of a PAMP by a Toll-like receptor. The structure of TLR3 bound to its PAMP, a fragment of double-stranded RNA, as seen from the side (top) and from above (bottom). Notice that the PAMP induces receptor dimerization by binding the surfaces on the side of each of the extracellular domains.
[Drawn from 3CIY.pdb].
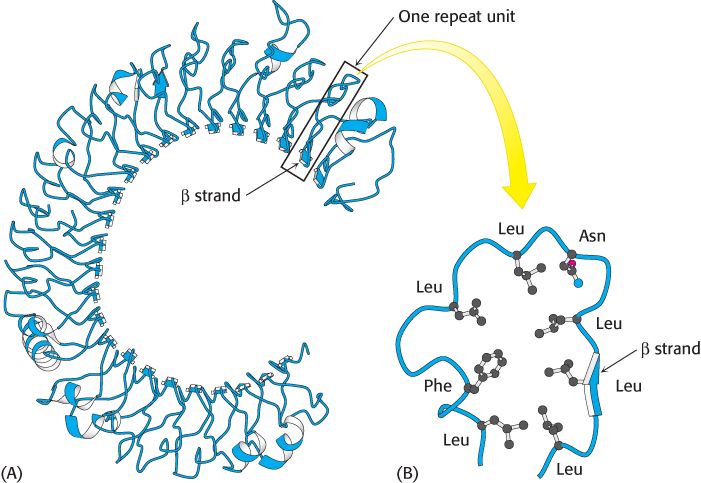
 FIGURE 34.2 Extracellular domain of the Toll-like receptor. (A) The structure of the leucine-rich repeat (LRR) domain from human TLR3. Notice that the LRR units come together to form a central parallel β sheet that curls to form a concave structure. (B) The structure of a single LRR showing the positions of the residues that are generally approximately conserved. Notice that the leucine residues come together to form a hydrophobic core with the single β strand on one side.
FIGURE 34.2 Extracellular domain of the Toll-like receptor. (A) The structure of the leucine-rich repeat (LRR) domain from human TLR3. Notice that the LRR units come together to form a central parallel β sheet that curls to form a concave structure. (B) The structure of a single LRR showing the positions of the residues that are generally approximately conserved. Notice that the leucine residues come together to form a hydrophobic core with the single β strand on one side.
[Drawn from 1ZIW.pdb].
Because the TLRs and other components of the innate immune system are always expressed, they provide the host organism with a rapid response to resist attack by pathogens. However, a number of pathogens have evolved the ability to escape detection by the innate immune system. For protection against such pathogens, the host relies on the adaptive immune system, which is able to target specific pathogens, even those that it has never encountered in the course of evolution.
The adaptive immune system responds by using the principles of evolution
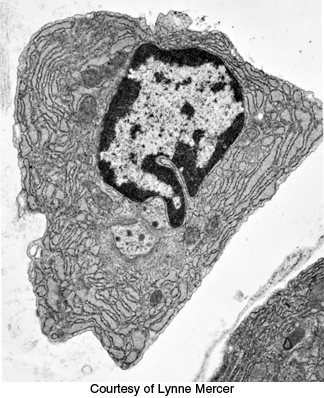
FIGURE 34.4Immunoglobulin production. An electron micrograph of a plasma cell shows the highly developed rough endoplasmic reticulum necessary for antibody secretion.
The adaptive immune system comprises two parallel but interrelated systems: humoral and cellular immune responses. In the humoral immune response, soluble proteins called antibodies (immunoglobulins) function as recognition elements that bind to foreign molecules and serve as markers signaling foreign invasion. Antibodies are secreted by plasma cells, which are derived from B lymphocytes (B cells) (Figure 34.4). A foreign macromolecule that binds selectively to an antibody is called an antigen. In a physiological context, if the binding of the foreign molecule stimulates an immune response, that molecule is called an immunogen. The specific affinity of an antibody is not for the entire macromolecular antigen but for a particular site on the antigen called the epitope or antigenic determinant. Each B cell produces just one type of antibody that can recognize a single epitope.
In the cellular immune response, cells called cytotoxic T lymphocytes (also commonly called killer T cells) destroy cells that have been invaded by a pathogen. Because intracellular pathogens do not leave markings on the exteriors of infected cells, vertebrates have evolved a mechanism to mark the exterior of cells with a sample of the interior contents, both self and foreign. Some of the internal proteins are broken into peptides, which are then bound to a complex of integral membrane proteins encoded by the major histocompatibility complex (MHC). T cells continually scan the bound peptides to find and kill cells that display foreign motifs on their surfaces. Another class of T cells called helper T lymphocytes (helper T cells) contributes to both the humoral and the cellular immune responses by stimulating the differentiation and proliferation of appropriate B cells and cytotoxic T cells. The cellular immune response is mediated by specific receptors that are expressed on the surfaces of the T cells.
The remarkable ability of the immune system to adapt to an essentially limitless set of potential pathogens requires a powerful system for transforming the immune cells and molecules in response to the presence of pathogens. This adaptive system operates through the principles of evolution, including reproduction with variation followed by selection of the most well suited members of a population.
If the human genome contains, by the latest estimates, only 21,000 genes, how can the immune system generate more than 108 different antibody proteins and 1012 T-cell receptors? The answer is found in a novel mechanism for generating a highly diverse set of genes from a limited set of genetic building blocks. Linking different sets of DNA regions in a combinatorial manner produces many distinct protein-encoding genes that are not present in the genome. A rigorous selection process then leaves for proliferation only cells that synthesize proteins determined to be useful in the immune response. The subsequent reproduction of these cells without additional recombination enriches the cell population with members expressing particular protein species.
Critical to the development of the immune response is the selection process, which determines which immune cells will reproduce. The process comprises several stages. In the early stages of immune cell development, cells expressing molecules that bind tightly to self-molecules are destroyed or silenced, whereas cells expressing molecules that do not bind strongly to self-molecules and that have the potential for binding strongly to foreign molecules are preserved. The appearance of an immunogenic invader at a later time will stimulate the reproduction of cells expressing antibodies or T-cell receptors that bind specifically to elements of that pathogen—in evolutionary terms, such cells are positively selected. Thus, the immune response is based on the selection of cells expressing molecules that are specifically effective against a particular invader; the response evolves from a population with wide-ranging specificities to a more-focused collection of cells and molecules that are well suited to defend the host when confronted with that particular challenge.
Not only are antibodies and T-cell receptors a result of genetic diversity and recombination, but antibodies have highly diverse structures as well. Antibodies require many different structural solutions for binding many different antigens, each of which has a different form. T-cell receptors, in contrast, are not structurally diverse, because they have coevolved with the MHC. The docking mode of a T-cell receptor to the peptide bound to MHC is similar for all structures. As a consequence of this coevolution, every T-cell receptor has an inherent reactivity with every MHC. The coevolution ensures that all T-cell receptors can scan all peptide–MHC complexes on all tissues. The genetic diversity of the 1012 different T-cell receptors is concentrated in a highly diverse set of residues in the center of the MHC groove. This localized diversity allows the T-cell receptor to recognize the many different foreign peptides bound to the MHC. T-cell receptors must survey many different MHC–peptide complexes with rapid turnover. Therefore, the binding affinities between T-cell receptors and the MHC are weaker than those between antibody and antigen.




 FIGURE 34.2 Extracellular domain of the Toll-
FIGURE 34.2 Extracellular domain of the Toll-