8.1Enzymes Are Powerful and Highly Specific Catalysts
Enzymes Are Powerful and Highly Specific Catalysts
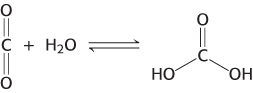
Enzymes accelerate reactions by factors of as much as a million or more (Table 8.1). Indeed, most reactions in biological systems do not take place at perceptible rates in the absence of enzymes. Even a reaction as simple as the hydration of carbon dioxide is catalyzed by an enzyme—
|
Enzyme |
Nonenzymatic half- |
Uncatalyzed rate (kun s−1) |
Catalyzed rate (kcat s−1) |
Rate enhancement (kcat s−1/kun s−1) |
|---|---|---|---|---|
|
OMP decarboxylase |
78,000,000 years |
2.8 × 10−16 |
39 |
1.4 × 1017 |
|
Staphylococcal nuclease |
130,000 years |
1.7 × 10−13 |
95 |
5.6 × 1014 |
|
AMP nucleosidase |
69,000 years |
1.0 × 10−11 |
60 |
6.0 × 1012 |
|
Carboxypeptidase A |
7.3 years |
3.0 × 10−9 |
578 |
1.9 × 1011 |
|
Ketosteroid isomerase |
7 weeks |
1.7 × 10−7 |
66,000 |
3.9 × 1011 |
|
Triose phosphate isomerase |
1.9 days |
4.3 × 10−6 |
4,300 |
1.0 × 109 |
|
Chorismate mutase |
7.4 hours |
2.6 × 10−5 |
50 |
1.9 × 106 |
|
Carbonic anhydrase |
5 seconds |
1.3 × 10−1 |
1 × 106 |
7.7 × 106 |
|
Abbreviations: OMP, orotidine monophosphate; AMP, adenosine monophosphate. Source: After A. Radzicka and R. Wolfenden. Science 267:90– |
||||
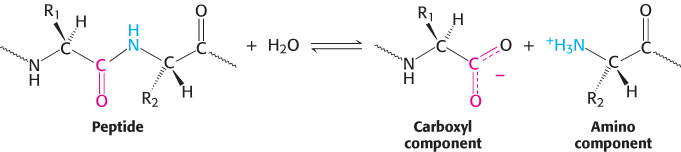
Enzymes are highly specific both in the reactions that they catalyze and in their choice of reactants, which are called substrates. An enzyme usually catalyzes a single chemical reaction or a set of closely related reactions. Let us consider proteolytic enzymes as an example. The biochemical function of these enzymes is to catalyze proteolysis, the hydrolysis of a peptide bond.
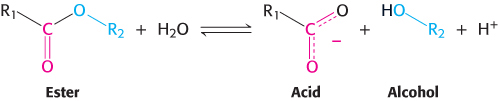
Most proteolytic enzymes also catalyze a different but related reaction in vitro—
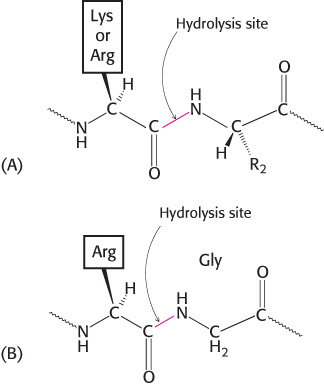
Proteolytic enzymes differ markedly in their degree of substrate specificity. Papain, which is found in papaya plants, is quite undiscriminating: it will cleave any peptide bond with little regard to the identity of the adjacent side chains. This lack of specificity accounts for its use in meat-
217
DNA polymerase I, a template-
Many enzymes require cofactors for activity
The catalytic activity of many enzymes depends on the presence of small molecules termed cofactors, although the precise role varies with the cofactor and the enzyme. Generally, these cofactors are able to execute chemical reactions that cannot be performed by the standard set of twenty amino acids. An enzyme without its cofactor is referred to as an apoenzyme; the complete, catalytically active enzyme is called a holoenzyme.
Apoenzyme + cofactor = holoenzyme
Cofactors can be subdivided into two groups: (1) metals and (2) small organic molecules called coenzymes (Table 8.2). Often derived from vitamins, coenzymes can be either tightly or loosely bound to the enzyme. Tightly bound coenzymes are called prosthetic groups. Loosely associated coenzymes are more like cosubstrates because, like substrates and products, they bind to the enzyme and are released from it. The use of the same coenzyme by a variety of enzymes sets coenzymes apart from normal substrates, however, as does their source in vitamins (Section 15.4). Enzymes that use the same coenzyme usually perform catalysis by similar mechanisms. In Chapter 9, we will examine the importance of metals to enzyme activity and, throughout the book, we will see how coenzymes and their enzyme partners operate in their biochemical context.
|
Cofactor |
Enzyme |
|---|---|
|
Coenzyme |
|
|
Thiamine pyrophosphate |
Pyruvate dehydrogenase |
|
Flavin adenine nucleotide |
Monoamine oxidase |
|
Nicotinamide adenine dinucleotide |
Lactate dehydrogenase |
|
Pyridoxal phosphate |
Glycogen phosphorylase |
|
Coenzyme A (CoA) |
Acetyl CoA carboxylase |
|
Biotin |
Pyruvate carboxylase |
|
5′-Deoxyadenosyl cobalamin |
Methylmalonyl mutase |
|
Tetrahydrofolate |
Thymidylate synthase |
|
Metal |
|
|
Zn2+ |
Carbonic anhydrase |
|
Zn2+ |
Carboxypeptidase |
|
Mg2+ |
EcoRV |
|
Mg2+ |
Hexokinase |
|
Ni2+ |
Urease |
|
Mo |
Nitrogenase |
|
Se |
Glutathione peroxidase |
|
Mn |
Superoxide dismutase |
|
K+ |
Acetyl CoA thiolase |
Enzymes can transform energy from one form into another
A key activity in all living systems is the conversion of one form of energy into another. For example, in photosynthesis, light energy is converted into chemical-
218
The molecular mechanisms of these energy-