
The impressive muscles of the horse, like the muscles of all animals, are powered by the molecular-motor protein myosin. A part of myosin moves dramatically (as shown above) in response to ATP binding, hydrolysis, and product release, propelling myosin along an actin filament. This molecular movement is translated into movement of the entire animal, vividly depicted in da Vinci’s rearing horse.
Organisms, from human beings to bacteria, move to adapt to changes in their environments, navigating toward food and away from danger. Cells themselves are not static but are bustling assemblies of moving proteins, nucleic acids, and organelles. This motion is enabled by two elements: molecular-motor proteins and complex networks of filamentous proteins termed the cytoskeleton (Figure 35.1). The dynamic networks that determine the shape and mobility of calls are among the most active areas of investigation in modern cell biology. Remarkably, the fundamental biochemical mechanisms that produce contractions in our muscles are the same as those that propel organelles along the cytoskeleton. In fact, many of the proteins that play key roles in converting chemical energy into kinetic energy are members of the same protein family, the P-loop NTPases, the hugely important group of proteins that we first examined in Chapter 9. These molecular motors are homologous to proteins that we have encountered in other contexts, including the G proteins in protein synthesis, signaling, and other processes. Once again, we see the economy of evolution in adapting existing proteins to perform new functions.
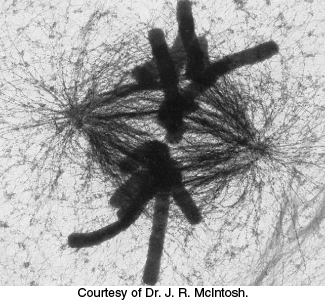
FIGURE 35.1Motion within cells. This high-voltage electron micrograph shows the mitotic apparatus in a metaphase mammalian cell. The large cylindrical objects are chromosomes, and the threadlike structures stretched across the center are microtubules, key components of the skeleton. Microtubules serve as tracks for the molecular motors that move chromosomes. Many processes, including chromosome segregation in mitosis, depend on the action of molecular-motor proteins.
Molecular motors operate by small increments, converting changes in protein conformation into directed motion. Orderly motion across distances requires a track that steers the motion of the motor assembly. Indeed, we have already encountered a class of molecular motors that utilize mechanisms that we will examine here—namely, the helicases that move along DNA during DNA replication (Section 28.1). The proteins on which we will focus in this chapter move along actin and microtubules—protein filaments composed of repeating subunits. The motor proteins cycle between forms having high or low affinity for the filament tracks in response to ATP binding and hydrolysis, enabling a bind, pull, and release mechanism that generates motion.
We will also consider a completely different strategy for generating motion, one used by bacteria such as E. coli. A set of flagella act as propellers, rotated by a motor in the bacterial cell membrane. This rotary motor is driven by a proton gradient across the membrane, rather than by ATP hydrolysis. The mechanism for coupling the proton gradient to rotatory motion is analogous to that used by the F0 subunit of ATP synthase. Thus, both of the major modes for storing biochemical energy—namely, ATP and ion gradients—have been harnessed by evolution to drive organized molecular motion.

