17.1The Pyruvate Dehydrogenase Complex Links Glycolysis to the Citric Acid Cycle
The Pyruvate Dehydrogenase Complex Links Glycolysis to the Citric Acid Cycle
Carbohydrates, most notably glucose, are processed by glycolysis into pyruvate (Chapter 16). Under anaerobic conditions, the pyruvate is converted into lactate or ethanol, depending on the organism. Under aerobic conditions, the pyruvate is transported into mitochondria by a specific carrier protein embedded in the mitochondrial membrane. In the mitochondrial matrix, pyruvate is oxidatively decarboxylated by the pyruvate dehydrogenase complex to form acetyl CoA.
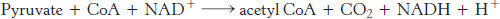
This irreversible reaction is the link between glycolysis and the citric acid cycle (Figure 17.4). Note that the pyruvate dehydrogenase complex produces CO2 and captures high-
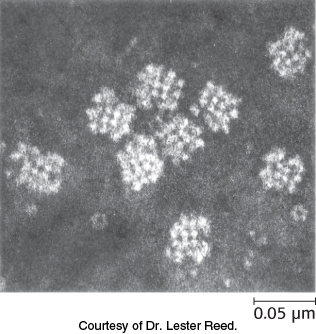
The pyruvate dehydrogenase complex is a large, highly integrated complex of three distinct enzymes (Table 17.1). Pyruvate dehydrogenase complex is a member of a family of homologous complexes that include the citric acid cycle enzyme α-ketoglutarate dehydrogenase complex. These complexes are giant, larger than ribosomes, with molecular masses ranging from 4 million to 10 million kDa (Figure 17.5). As we will see, their elaborate structures allow groups to travel from one active site to another, connected by tethers to the core of the structure.
|
Enzyme |
Abbreviation |
Number of chains |
Prosthetic group |
Reaction catalyzed |
|---|---|---|---|---|
|
Pyruvate dehydrogenase component |
E1 |
24 |
TPP |
Oxidative decarboxylation of pyruvate |
|
Dihydrolipoyl transacetylase |
E2 |
24 |
Lipoamide |
Transfer of acetyl group to CoA |
|
Dihydrolipoyl dehydrogenase |
E3 |
12 |
FAD |
Regeneration of the oxidized form of lipoamide |
498
Mechanism: The synthesis of acetyl coenzyme A from pyruvate requires three enzymes and five coenzymes
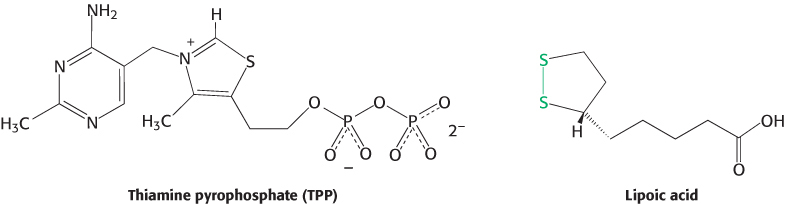
The mechanism of the pyruvate dehydrogenase reaction is wonderfully complex, more so than is suggested by its simple stoichiometry. The reaction requires the participation of the three enzymes of the pyruvate dehydrogenase complex and five coenzymes. The coenzymes thiamine pyrophosphate (TPP), lipoic acid, and FAD serve as catalytic cofactors, and CoA and NAD+ are stoichiometric cofactors, cofactors that function as substrates.
The conversion of pyruvate into acetyl CoA consists of three steps: decarboxylation, oxidation, and transfer of the resultant acetyl group to CoA. A fourth step is required to regenerate the active enzyme.
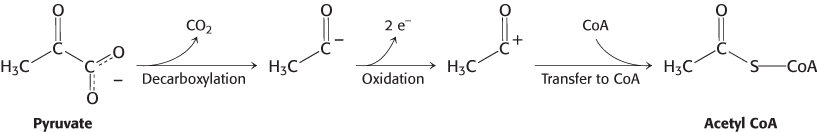
These steps must be coupled to preserve the free energy derived from the decarboxylation step to drive the formation of NADH and acetyl CoA:
Decarboxylation. Pyruvate combines with TPP and is then decarboxylated to yield hydroxyethyl-
TPP (Figure 17.6). 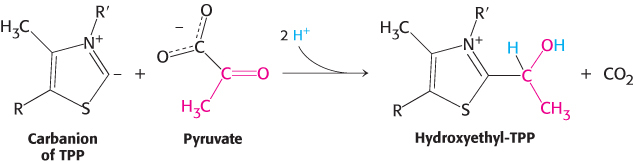
This reaction, the rate limiting step in acetyl CoA synthesis, is catalyzed by the pyruvate dehydrogenase component (E1) of the multienzyme complex. TPP is the prosthetic group of the pyruvate dehydrogenase component.
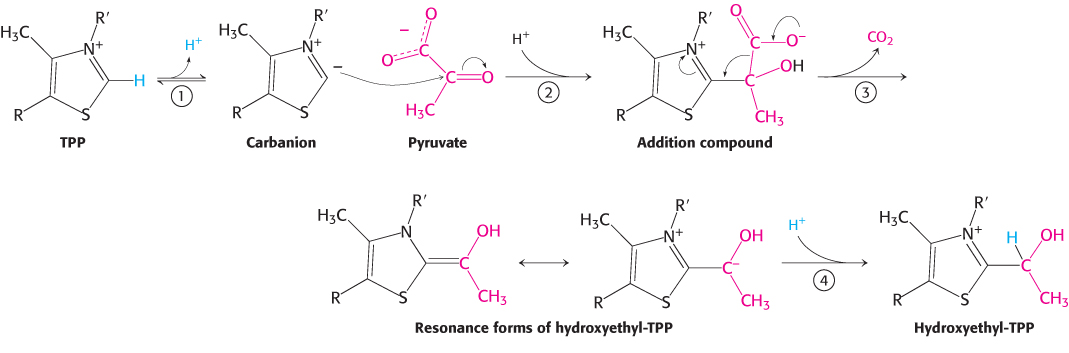 FIGURE 17.6Mechanism of the E1 decarboxylation reaction. E1 is the pyruvate dehydrogenase component of the pyruvate dehydrogenase complex. A key feature of the prosthetic group, TPP, is that the carbon atom between the nitrogen and sulfur atoms in the thiazole ring is much more acidic than most =C—
FIGURE 17.6Mechanism of the E1 decarboxylation reaction. E1 is the pyruvate dehydrogenase component of the pyruvate dehydrogenase complex. A key feature of the prosthetic group, TPP, is that the carbon atom between the nitrogen and sulfur atoms in the thiazole ring is much more acidic than most =C—groups, with a pKa value near 10. (1) This carbon center ionizes to form a carbanion. (2) The carbanion readily adds to the carbonyl group of pyruvate. (3) This addition is followed by the decarboxylation of pyruvate. The positively charged ring of TPP acts as an electron sink that stabilizes the negative charge that is transferred to the ring as part of the decarboxylation. (4) Protonation yields hydroxyethyl- TPP. Oxidation. The hydroxyethyl group attached to TPP is oxidized to form an acetyl group while being simultaneously transferred to lipoamide, a derivative of lipoic acid that is linked to the side chain of a lysine residue by an amide linkage. Note that this transfer results in the formation of an energy-
rich thioester bond. 499
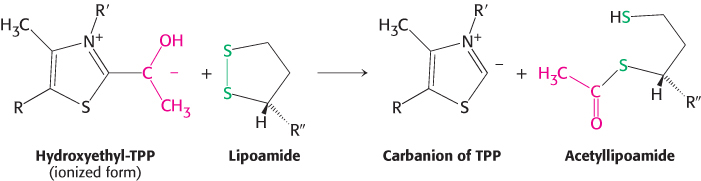
The oxidant in this reaction is the disulfide group of lipoamide, which is reduced to its disulfhydryl form. This reaction, also catalyzed by the pyruvate dehydrogenase component E1, yields acetyllipoamide.
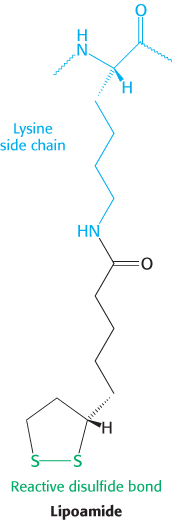
Formation of Acetyl CoA. The acetyl group is transferred from acetyllipoamide to CoA to form acetyl CoA.
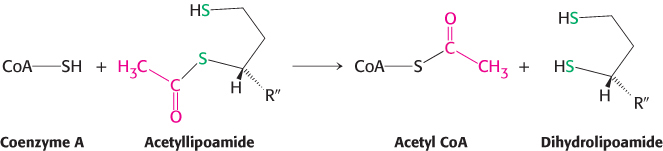
Dihydrolipoyl transacetylase (E2) catalyzes this reaction. The energy-
rich thioester bond is preserved as the acetyl group is transferred to CoA. Recall that CoA serves as a carrier of many activated acyl groups, of which acetyl is the simplest (Section 15.3). Acetyl CoA, the fuel for the citric acid cycle, has now been generated from pyruvate. Regeneration of oxidized lipoamide. The pyruvate dehydrogenase complex cannot complete another catalytic cycle until the dihydrolipoamide is oxidized to lipoamide. In the fourth step, the oxidized form of lipoamide is regenerated by dihydrolipoyl dehydrogenase (E3). Two electrons are transferred to an FAD prosthetic group of the enzyme and then to NAD+.
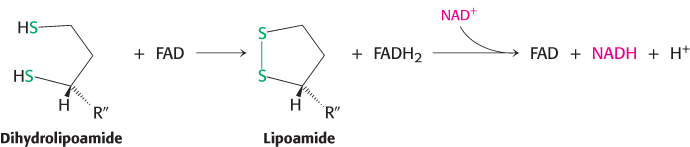
500
This electron transfer from FAD to NAD+ is unusual because the common role for FAD is to receive electrons from NADH. The electron-
Flexible linkages allow lipoamide to move between different active sites
The structures of all of the component enzymes of the bacterial pyruvate dehydrogenase complex are known, albeit from different complexes and species. Thus, it is now possible to construct an atomic model of the complex to understand its activity (Figure 17.7).
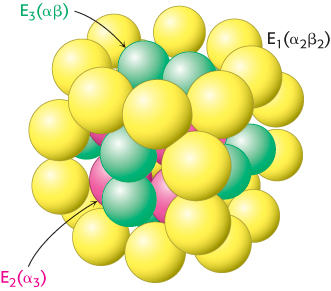
The core of the complex is formed by the transacetylase component E2. Transacetylase consists of eight catalytic trimers assembled to form a hollow cube. Each of the three subunits forming a trimer has three major domains (Figure 17.8). At the amino terminus is a small domain that contains a bound flexible lipoamide cofactor attached to a lysine residue. This domain is homologous to biotin-
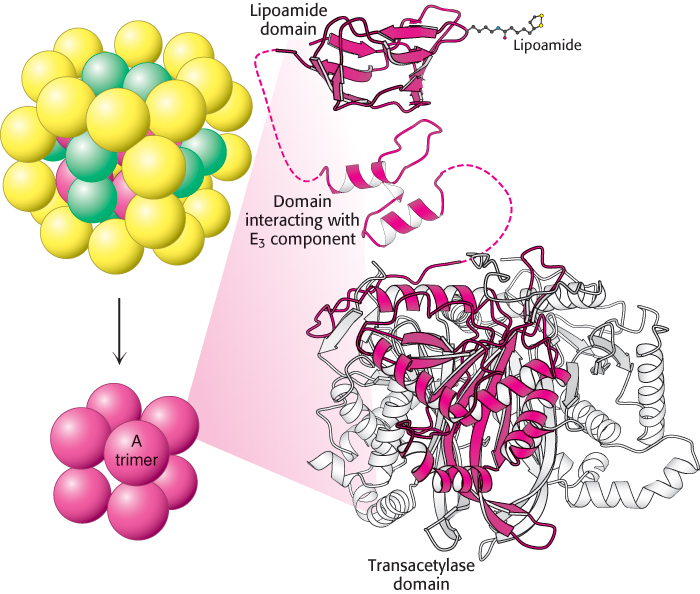
How do the three distinct active sites work in concert (Figure 17.9)? The key is the long, flexible lipoamide arm of the E2 subunit, which carries substrate from active site to active site:
Pyruvate is decarboxylated at the active site of E1, forming the hydroxyethyl-
TPP intermediate, and CO2 leaves as the first product. This active site lies deep within the E1 complex, connected to the enzyme surface by a 20- Å-long hydrophobic channel. 501
E2 inserts the lipoamide arm of the lipoamide domain into the deep channel in E1 leading to the active site.
E1 catalyzes the transfer of the acetyl group to the lipoamide. The acetylated arm then leaves E1 and enters the E2 cube to visit the active site of E2, located deep in the cube at the subunit interface.
The acetyl moiety is then transferred to CoA, and the second product, acetyl CoA, leaves the cube. The reduced lipoamide arm then swings to the active site of the E3 flavoprotein.
At the E3 active site, the lipoamide is oxidized by coenzyme FAD. The reactivated lipoamide is ready to begin another reaction cycle.
The final product, NADH, is produced with the reoxidation of FADH2 to FAD.
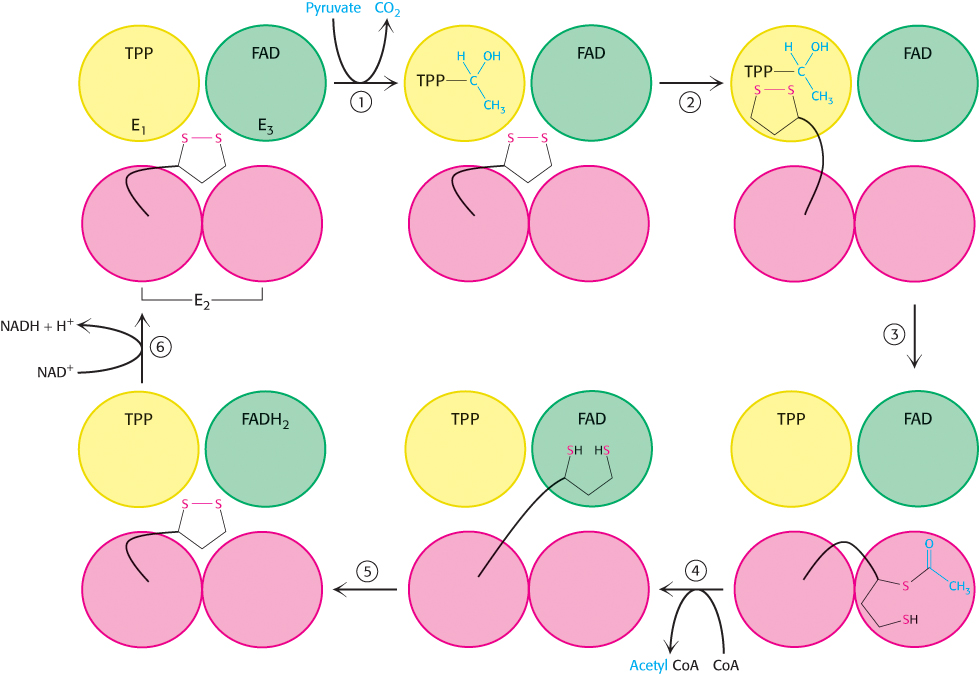
The structural integration of three kinds of enzymes and the long, flexible lipoamide arm make the coordinated catalysis of a complex reaction possible. The proximity of one enzyme to another increases the overall reaction rate and minimizes side reactions. All the intermediates in the oxidative decarboxylation of pyruvate remain bound to the complex throughout the reaction sequence and are readily transferred as the flexible arm of E2 calls on each active site in turn.