18.4
A Proton Gradient Powers the Synthesis of ATP
Thus far, we have considered the flow of electrons from NADH to O2, an exergonic process.
Next, we consider how this process is coupled to the synthesis of ATP, an endergonic process.
A molecular assembly in the inner mitochondrial membrane carries out the synthesis of ATP. This enzyme complex was originally called the mitochondrial ATPase or F1F0 ATPase because it was discovered through its catalysis of the reverse reaction, the hydrolysis of ATP. ATP synthase, its preferred name, emphasizes its actual role in the mitochondrion. It is also called Complex V.
How is the oxidation of NADH coupled to the phosphorylation of ADP? Electron transfer was first suggested to lead to the formation of a covalent high-energy intermediate that serves as a compound having a high phosphoryl-transfer potential, analogous to the generation of ATP by the formation of 1,3-bisphosphoglycerate in glycolysis (Section 16.1). An alternative proposal was that electron transfer aids the formation of an activated protein conformation, which then drives ATP synthesis. The search for such intermediates for several decades proved fruitless.
Some have argued that, along with the elucidation of the structure of DNA, the discovery that ATP synthesis is powered by a proton gradient is one of the two major advances in biology in the twentieth century. Mitchell’s initial postulation of the chemiosmotic theory was not warmly received by all. Efraim Racker, one of the early investigators of ATP synthase, recalls that some thought of Mitchell as a court jester, whose work was of no consequence. Peter Mitchell was awarded the Nobel Prize in chemistry in 1978 for his contributions to understanding oxidative phosphorylation.
In 1961, Peter Mitchell suggested a radically different mechanism, the chemiosmotic hypothesis. He proposed that electron transport and ATP synthesis are coupled by a proton gradient across the inner mitochondrial membrane. In his model, the transfer of electrons through the respiratory chain leads to the pumping of protons from the matrix to the cytoplasmic side of the inner mitochondrial membrane. The H+ concentration becomes lower in the matrix, and an electric field with the matrix side negative is generated (Figure 18.22). Protons then flow back into the matrix to equalize the distribution. Mitchell’s idea was that this flow of protons drives the synthesis of ATP by ATP synthase. The energy-rich unequal distribution of protons is called the proton-motive force. The proton-motive force is composed of two components: a chemical gradient and a charge gradient. The chemical gradient for protons can be represented as a pH gradient. The charge gradient is created by the positive charge on the unequally distributed protons forming the chemical gradient. Mitchell proposed that both components power the synthesis of ATP.
Proton-motive force(Δp) = chemical gradient(ΔpH) + charge gradient(Δψ)
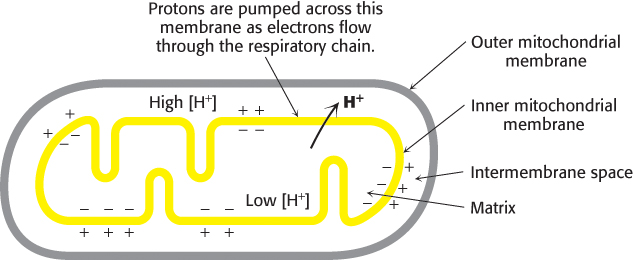
FIGURE 18.22Chemiosmotic hypothesis. Electron transfer through the respiratory chain leads to the pumping of protons from the matrix to the cytoplasmic side of the inner mitochondrial membrane. The pH gradient and membrane potential constitute a proton-motive force that is used to drive ATP synthesis.
Mitchell’s highly innovative hypothesis that oxidation and phosphorylation are coupled by a proton gradient is now supported by a wealth of evidence. Indeed, electron transport does generate a proton gradient across the inner mitochondrial membrane. The pH outside is 1.4 units lower than inside, and the membrane potential is 0.14 V, the outside being positive. As calculated on page 529, this membrane potential corresponds to a free energy of 21.8 kJ (5.2 kcal) per mole of protons.
An artificial system was created to elegantly demonstrate the basic principle of the chemiosmotic hypothesis. The role of the respiratory chain was played by bacteriorhodopsin, a membrane protein from halobacteria that pumps protons when illuminated. Synthetic vesicles containing bacteriorhodopsin and mitochondrial ATP synthase purified from beef heart were created (Figure 18.23). When the vesicles were exposed to light, ATP was formed. This key experiment clearly showed that the respiratory chain and ATP synthase are biochemically separate systems, linked only by a proton-motive force.
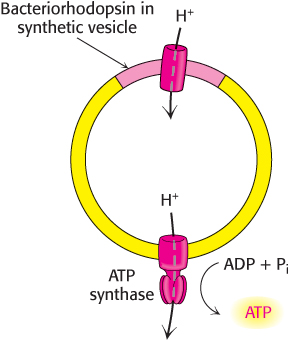
FIGURE 18.23Testing the chemiosmotic hypothesis. ATP is synthesized when reconstituted membrane vesicles containing bacteriorhodopsin (a light-driven proton pump) and ATP synthase are illuminated.
ATP synthase is composed of a proton-conducting unit and a catalytic unit
Two parts of the puzzle of how NADH oxidation is coupled to ATP synthesis are now evident: (1) electron transport generates a proton-motive force, and (2) ATP synthesis by ATP synthase can be powered by a proton-motive force. How is the proton-motive force converted into the high phosphoryl-transfer potential of ATP?
Biochemical, electron microscopic, and crystallographic studies of ATP synthase have revealed many details of its structure (Figure 18.24). It is a large, complex enzyme resembling a ball on a stick. Much of the “stick” part, called the F0 subunit, is embedded in the inner mitochondrial membrane. The 85-Å-diameter ball, called the F1 subunit, protrudes into the mitochondrial matrix. The F1 subunit contains the catalytic activity of the synthase. In fact, isolated F1 subunits display ATPase activity.
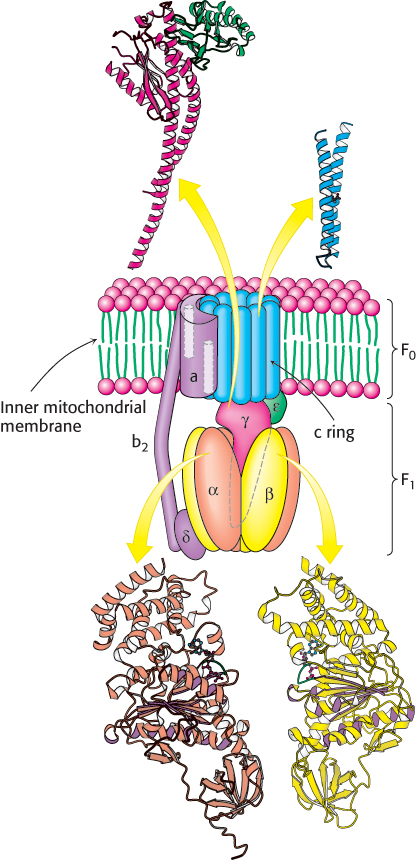
 FIGURE 18.24 Structure of ATP synthase. A schematic structure is shown along with representations of the components for which structures have been determined to high resolution. The P-loop NTPase domains of the α and β subunits are indicated by purple shading. Notice that part of the enzyme complex is embedded in the inner mitochondrial membrane, whereas the remainder resides in the matrix.
FIGURE 18.24 Structure of ATP synthase. A schematic structure is shown along with representations of the components for which structures have been determined to high resolution. The P-loop NTPase domains of the α and β subunits are indicated by purple shading. Notice that part of the enzyme complex is embedded in the inner mitochondrial membrane, whereas the remainder resides in the matrix.
[Drawn from 1E79.pdb and 1C0V.pdb.]
The F1 subunit consists of five types of polypeptide chains (α3, β3, γ, δ, and ε) with the indicated stoichiometry. The α and β subunits, which make up the bulk of the F1, are arranged alternately in a hexameric ring; they are homologous to one another and are members of the P-loop NTPase family (Section 9.4). Both bind nucleotides but only the β subunits are catalytically active. Just below the α and β subunits is a central stalk consisting of the γ and ε proteins. The γ subunit includes a long helical coiled coil that extends into the center of the α3β3 hexamer. The γ subunit breaks the symmetry of the α3β3 hexamer: each of the γ subunits is distinct by virtue of its interaction with a different face of γ. Distinguishing the three β subunits is crucial for understanding the mechanism of ATP synthesis.
The F0 subunit is a hydrophobic segment that spans the inner mitochondrial membrane. F0 contains the proton channel of the complex. This channel consists of a ring comprising from 8 to 14 c subunits that are embedded in the membrane. A single a subunit binds to the outside of the ring. The F0 and F1 subunits are connected in two ways: by the central γε stalk and by an exterior column. The exterior column consists of one a subunit, two b subunits, and the δ subunit.
ATP synthases associate with one another to form dimers, which then associate to form large oligomers of dimers. This association stabilizes the individual enzymes to the rotational forces required for catalysis and facilitates the curvature of the inner mitochondrial membrane. The formation of the cristae allows the proton pumps of the electron transport chain to localize the proton gradient in the vicinity of the synthases, which are located at the tips of the cristae, thereby enhancing the efficiency of ATP synthesis (Figure 18.25).
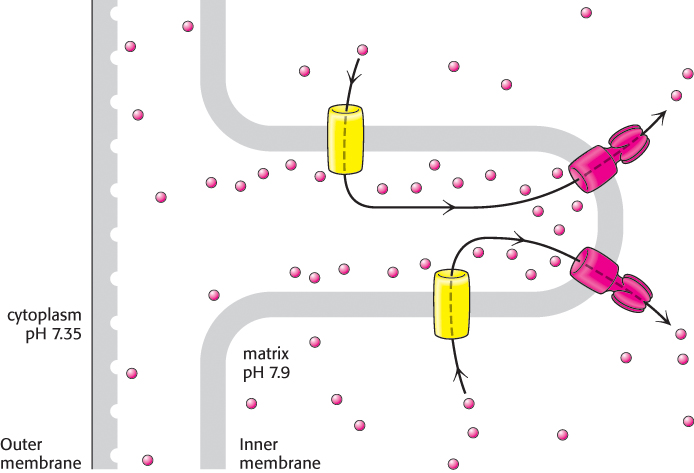
FIGURE 18.25ATPase assists in the formation of cristae. Oligomers of ATP synthase dimers facilitate cristae formation, creating an area where the protons (red balls) are concentrated and have ready access to the Fo portion of the ATP synthase. The electron transport chain is represented by the yellow cylinders embedded in the inner mitochondrial membrane.
[Information from K. M. Davies et al., Proc. Natl. Acad. Sci. U.S.A. 108: 14121–14126, 2011.]
Proton flow through ATP synthase leads to the release of tightly bound ATP: The binding-change mechanism
ATP synthase catalyzes the formation of ATP from ADP and orthophosphate.
The actual substrates are ADP and ATP complexed with Mg2+, as in all known phosphoryl-transfer reactions with these nucleotides. A terminal oxygen atom of ADP attacks the phosphorus atom of Pi to form a pentacovalent intermediate, which then dissociates into ATP and H2O (Figure 18.26).
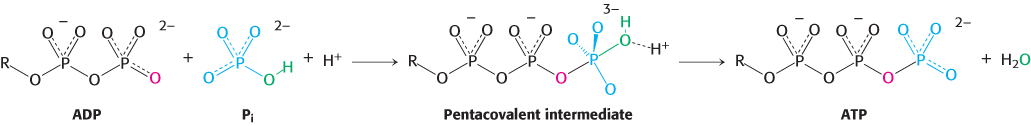
FIGURE 18.26ATP-synthesis mechanism. One of the oxygen atoms of ADP attacks the phosphorus atom of Pi to form a pentacovalent intermediate, which then forms ATP and releases a molecule of H2O.
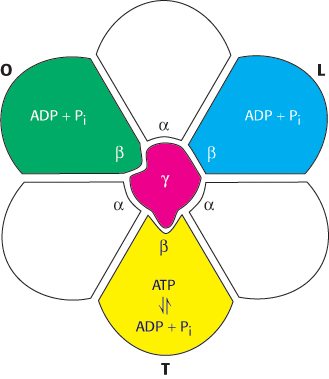
FIGURE 18.28ATP synthase nucleotide-binding sites are not equivalent. The γ subunit passes through the center of the α3β3 hexamer and makes the nucleotide-binding sites in the β subunits distinct from one another. The β subunits are colored to distinguish them from one another.
How does the flow of protons drive the synthesis of ATP? Isotopic-exchange experiments unexpectedly revealed that enzyme-bound ATP forms readily in the absence of a proton-motive force. When ADP and Pi were added to ATP synthase in H218O,18O became incorporated into Pi through the synthesis of ATP and its subsequent hydrolysis (Figure 18.27). The rate of incorporation of18O into Pi showed that about equal amounts of bound ATP and ADP are in equilibrium at the catalytic site, even in the absence of a proton gradient. However, ATP does not leave the catalytic site unless protons flow through the enzyme. Thus, the role of the proton gradient is not to form ATP but to release it from the synthase.
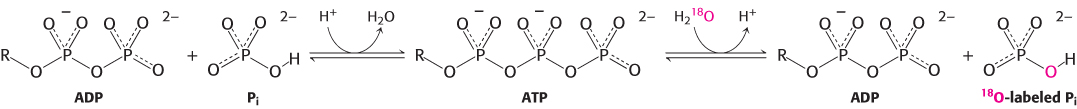
FIGURE 18.27ATP forms without a proton-motive force but is not released. The results of isotopic-exchange experiments indicate that enzyme-bound ATP is formed from ADP and Pi in the absence of a proton-motive force.
The fact that three β subunits are components of the F1 moiety of the ATPase means that there are three active sites on the enzyme, each performing one of three different functions at any instant. The proton-motive force causes the three active sites to sequentially change functions as protons flow through the membrane-embedded component of the enzyme. Indeed, we can think of the enzyme as consisting of a moving part and a stationary part: (1) the moving unit, or rotor, consists of the c ring and the γϵ stalk and (2) the stationary unit, or stator, is composed of the remainder of the molecule.
How do the three active sites of ATP synthase respond to the flow of protons? A number of experimental observations suggested a binding-change mechanism for proton-driven ATP synthesis. This proposal states that a β subunit can perform each of three sequential steps in the synthesis of ATP by changing conformation. These steps are (1) ADP and Pi binding, (2) ATP synthesis, and (3) ATP release. As already noted, interactions with the γ subunit make the three β subunits structurally distinct (Figure 18.28). At any given moment, one β subunit will be in the L, or loose, conformation. This conformation binds ADP and Pi. A second subunit will be in the T, or tight, conformation. This conformation binds ATP with great avidity, so much so that it will convert bound ADP and Pi into ATP. Both the T and L conformations are sufficiently constrained that they cannot release bound nucleotides. The final subunit will be in the O, or open, form. This form has a more open conformation and can bind or release adenine nucleotides.
Progressive alteration of the forms of the three active sites of ATP synthase
The rotation of the γ subunit drives the interconversion of these three forms (Figure 18.29). ADP and Pi bound in the subunit in the T form are transiently combining to form ATP. Suppose that the γ subunit is rotated by 120 degrees in a counterclockwise direction (as viewed from the top). This rotation converts the T-form site into an O-form site with the nucleotide bound as ATP. Concomitantly, the L-form site is converted into a T-form site, enabling the transformation of an additional ADP and Pi into ATP. The ATP in the O-form site can now depart from the enzyme to be replaced by ADP and Pi. An additional 120-degree rotation converts this O-form site into an L-form site, trapping these substrates. Each subunit progresses from the T to the O to the L form with no two subunits ever present in the same conformational form. This mechanism suggests that ATP can be synthesized and released by driving the rotation of the γ subunit in the appropriate direction.
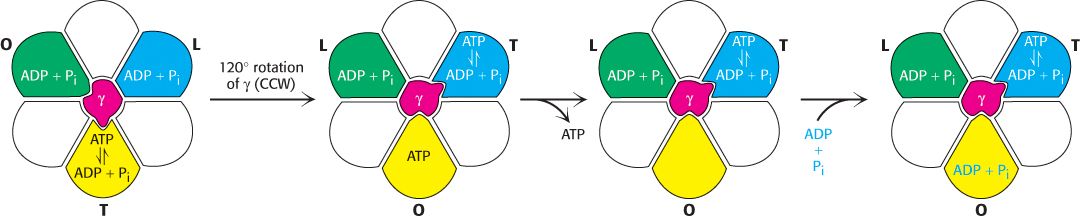
FIGURE 18.29Binding-change mechanism for ATP synthase. The rotation of the γ subunit interconverts the three β subunits. The subunit in the T (tight) form interconverts ADP and Pi and ATP but does not allow ATP to be released. When the γ subunit is rotated by 120 degrees in a counterclockwise (CCW) direction, the T-form subunit is converted into the O form, allowing ATP release. ADP and Pi can then bind to the O-form subunit. An additional 120-degree rotation (not shown) traps these substrates in an L-form subunit.
Rotational catalysis is the world’s smallest molecular motor
Is it possible to observe the proposed rotation directly? Elegant experiments, using single-molecule techniques (Section 8.6), have demonstrated the rotation through the use of a simple experimental system consisting solely of cloned α3β3γ subunits (Figure 18.30). The β subunits were engineered to contain amino-terminal polyhistidine tags, which have a high affinity for nickel ions (Section 3.1). This property of the tags allowed the α3β3 assembly to be immobilized on a glass surface that had been coated with nickel ions. The γ subunit was linked to a fluorescently labeled actin filament to provide a long segment that could be observed under a fluorescence microscope. Remarkably, the addition of ATP caused the actin filament to rotate unidirectionally in a counterclockwise direction. The γ subunit was rotating, driven by the hydrolysis of ATP. Thus, the catalytic activity of an individual molecule could be observed. The counterclockwise rotation is consistent with the predicted mechanism for hydrolysis because the molecule was viewed from below relative to the view shown in Figure 18.30.
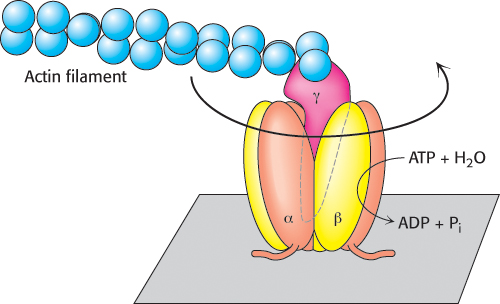
FIGURE 18.30Direct observation of ATP-driven rotation in ATP synthase. The α3β3 hexamer of ATP synthase is fixed to a surface, with the γ subunit projecting upward and linked to a fluorescently labeled actin filament. The addition and subsequent hydrolysis of ATP result in the counterclockwise rotation of the γ subunit, which can be directly seen under a fluorescence microscope.
More-detailed analysis in the presence of lower concentrations of ATP revealed that the γ subunit rotates in 120-degree increments. Each increment corresponds to the hydrolysis of a single ATP molecule. In addition, from the results obtained by varying the length of the actin filament and measuring the rate of rotation, the enzyme appears to operate near 100% efficiency; that is, essentially all of the energy released by ATP hydrolysis is converted into rotational motion.
Proton flow around the c ring powers ATP synthesis
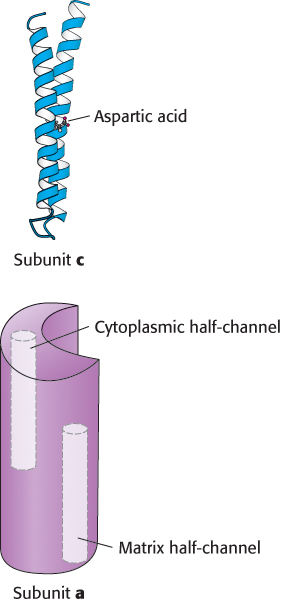
FIGURE 18.31Components of the proton-conducting unit of ATP synthase. The c subunit consists of two α helices that span the membrane. In E. coli, an aspartic acid residue in one of the helices lies on the center of the membrane. The structure of the a subunit has not yet been directly observed, but it appears to include two half-channels that allow protons to enter and pass partway but not completely through the membrane.
The direct observation of rotary motion of the γ subunit is strong evidence for the rotational mechanism for ATP synthesis. The last remaining question is: How does proton flow through F0 drive the rotation of the γ subunit? The mechanism depends on the structures of the a and c subunits of F0 (Figure 18.31). The stationary a subunit directly abuts the membrane-spanning ring formed by 8 to 14 c subunits. The a subunit includes two hydrophilic half-channels that do not span the membrane (Figure 18.31). Thus, protons can pass into either of these channels, but they cannot move completely across the membrane. The a subunit is positioned such that each half-channel directly interacts with one c subunit.
The structure of the c subunit was determined both by NMR methods and by x-ray crystallography. Each polypeptide chain forms a pair of α helices that span the membrane. A glutamic acid (or aspartic acid) residue is found in the middle of one of the helices. If the glutamate is charged (unprotonated), the c subunit will not move into the membrane. The key to proton movement across the membrane is that, in a proton-rich environment, such as the cytoplasmic side of the inner mitochondrial membrane, a proton will enter a channel and bind the glutamate residue, while the glutamic acid in a proton-poor environment of the other half-channel will release a proton (Figure 18.32). The c subunit with the bound proton then moves into the membrane as the ring rotates by one c subunit. This rotation brings the newly deprotonated c subunit from the matrix half-channel to the proton rich cytoplasmic half-channel, where it can bind a proton. The movement of protons through the half-channels from the high proton concentration of the cytoplasm to the low proton concentration of the matrix powers the rotation of the c ring. The a unit remains stationary as the c ring rotates. Each proton that enters the cytoplasmic half-channel of the a unit moves through the membrane by riding around on the rotating c ring to exit through the matrix half-channel into the proton-poor environment of the matrix (Figure 18.33).
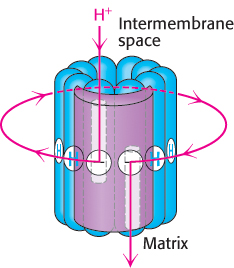
FIGURE 18.33Proton path through the membrane. Each proton enters the cytoplasmic half-channel, follows a complete rotation of the c ring, and exits through the other half-channel into the matrix.
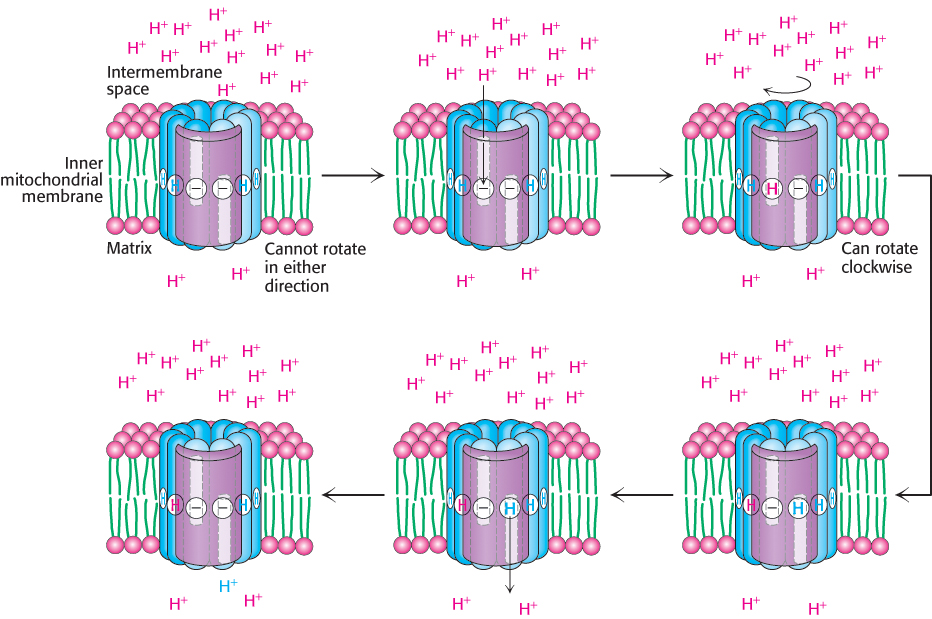
FIGURE 18.32Proton motion across the membrane drives rotation of the c ring. A proton enters from the intermembrane space into the cytoplasmic half-channel to neutralize the charge on an aspartate residue in a c subunit. With this charge neutralized, the c ring can rotate clockwise by one c subunit, moving an aspartic acid residue out of the membrane into the matrix half-channel. This proton can move into the matrix, resetting the system to its initial state.
How does the rotation of the c ring lead to the synthesis of ATP? The c ring is tightly linked to the γ and ε subunits. Thus, as the c ring turns, the γ and ε subunits are turned inside the α3β3 hexamer unit of F1. The rotation of the γ subunit in turn promotes the synthesis of ATP through the binding-change mechanism. The exterior column formed by the two b chains and the δ subunit prevents the α3β3 hexamer from rotating in sympathy with the c ring. Recall that the number of c subunits in the c ring appears to range between 8 and 14. This number is significant because it determines the number of protons that must be transported to generate a molecule of ATP. Each 360-degree rotation of the γ subunit leads to the synthesis and release of three molecules of ATP. Thus, if there are 10 c subunits in the ring (as was observed in a crystal structure of yeast mitochondrial ATP synthase), each ATP generated requires the transport of 10/3 = 3.33 protons. Recent evidence shows that the c rings of all vertebrates are composed of 8 subunits, making vertebrate ATP synthase the most-efficient ATP synthase known, with the transport of only 2.7 protons required for ATP synthesis. For simplicity, we will assume that three protons must flow into the matrix for each ATP formed, but we must keep in mind that the true value may differ. As we will see, the electrons from NADH pump enough protons to generate 2.5 molecules of ATP, whereas those from FADH2 yield 1.5 molecules of ATP.
A little goes a long way
Despite the various molecular machinations and the vast numbers of ATPs synthesized and protons pumped, a resting human being requires surprisingly little power. Approximately 116 watts, the energy output of a typical light bulb, provides enough energy to sustain a resting person.
Let us return for a moment to the example with which we began this chapter. If a resting human being requires 85 kg of ATP per day for bodily functions, then 3.3 × 1025 protons must flow through the ATP synthase per day, or 3.3 × 1021 protons per second. Figure 18.34 summarizes the process of oxidative phosphorylation.
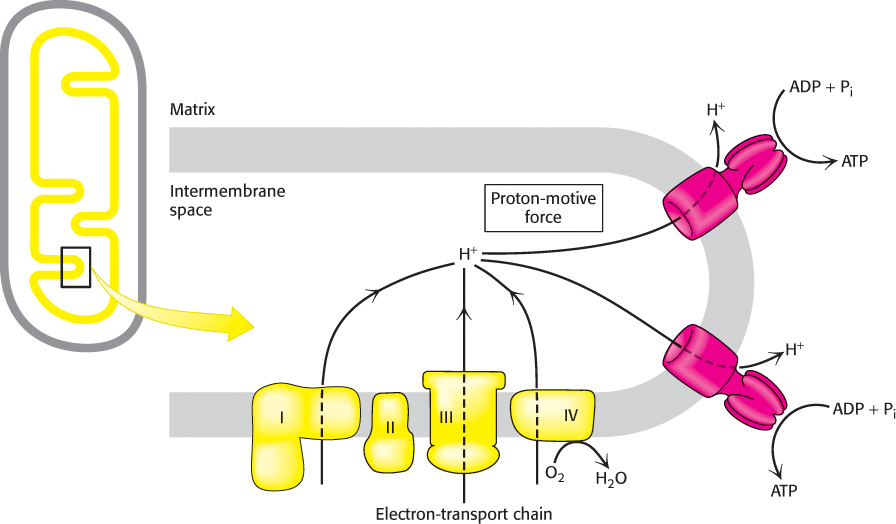
FIGURE 18.34Overview of oxidative phosphorylation. The electron-transport chain generates a proton gradient, which is used to synthesize ATP.
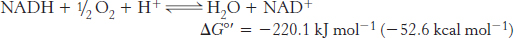
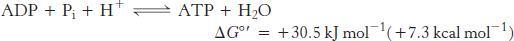



 FIGURE 18.24 Structure of ATP synthase. A schematic structure is shown along with representations of the components for which structures have been determined to high resolution. The P-
FIGURE 18.24 Structure of ATP synthase. A schematic structure is shown along with representations of the components for which structures have been determined to high resolution. The P-
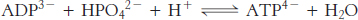



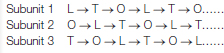






 The α and β subunits of ATP synthase are members of the P-
The α and β subunits of ATP synthase are members of the P-