30.3
The Ribosome Is the Site of Protein Synthesis
We turn now to ribosomes, the molecular machines that coordinate the interplay of aminoacyl-tRNAs, mRNA, and proteins that leads to protein synthesis. An E. coli ribosome is a ribonucleoprotein assembly with a mass of about 2500 kDa, a diameter of approximately 250 Å, and a sedimentation coefficient (Section 3.1) of 70S. The 20,000 ribosomes in a bacterial cell constitute nearly a fourth of its mass.
A ribosome can be dissociated into a large subunit (50S) and a small subunit (30S). These subunits can be further split into their constituent proteins and RNAs. The 30S subunit contains 21 different proteins (referred to as S1 through S21) and a 16S RNA molecule. The 50S subunit contains 34 different proteins (L1 through L34) and two RNA molecules, a 23S and a 5S species. A ribosome contains one copy of each RNA molecule, two copies each of the L7 and L12 proteins, and one copy of each of the other proteins. The L7 protein is identical with L12 except that its amino terminus is acetylated (Section 10.3). Both the 30S and the 50S subunits can be reconstituted in vitro from their constituent proteins and RNA. This reconstitution is an outstanding example of the principle that supramolecular complexes can form spontaneously from their macromolecular constituents.
The structures of both the 30S and the 50S subunits as well as the complete 70S ribosome have been determined (Figure 30.14). The features of these structures are in remarkable agreement with interpretations of less-direct experimental probes. These structures provide an invaluable framework for examining the mechanism of protein synthesis.
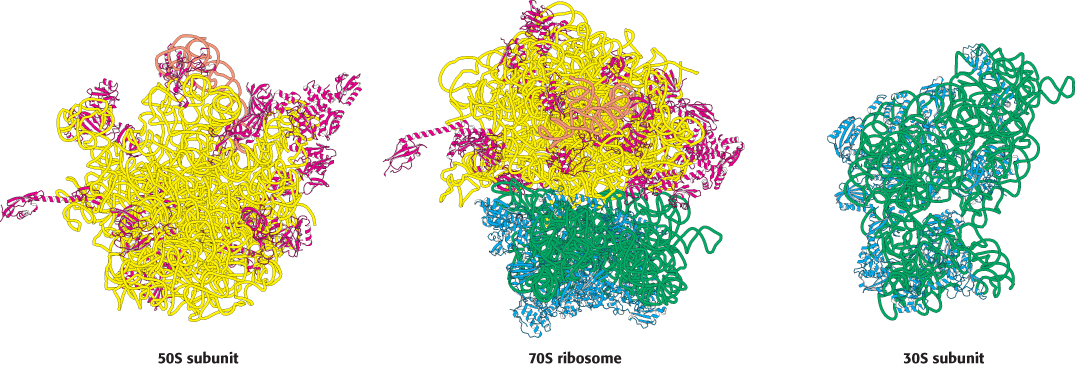
 FIGURE 30.14 The ribosome at high resolution. Detailed models of the ribosome based on the results of x-ray crystallographic studies of the 70S ribosome and the 30S and 50S subunits: (left) view of the part of the 50S subunit that interacts with the 30S subunit; (center) side view of the 70S ribosome; (right) view of the part of the 30S subunit that interacts with the 50S subunit. 23S RNA is shown in yellow, 5S RNA in orange, 16S RNA in green, proteins of the 50S subunit in red, and proteins of the 30S subunit in blue. Notice that the interface between the 50S and the 30S subunits consists entirely of RNA.
FIGURE 30.14 The ribosome at high resolution. Detailed models of the ribosome based on the results of x-ray crystallographic studies of the 70S ribosome and the 30S and 50S subunits: (left) view of the part of the 50S subunit that interacts with the 30S subunit; (center) side view of the 70S ribosome; (right) view of the part of the 30S subunit that interacts with the 50S subunit. 23S RNA is shown in yellow, 5S RNA in orange, 16S RNA in green, proteins of the 50S subunit in red, and proteins of the 30S subunit in blue. Notice that the interface between the 50S and the 30S subunits consists entirely of RNA.
[Drawn from 1GIX. pdb and 1GIY.pdb.]
Ribosomal RNAs (5S, 16S, and 23S rRNA) play a central role in protein synthesis
The prefix ribo in the name ribosome is apt because RNA constitutes nearly two-thirds of the mass of these large molecular assemblies. The three RNAs present—5S, 16S, and 23S—are critical for ribosomal architecture and function. They are formed by the cleavage of primary 30S transcripts and further processing. These molecules fold into structures that allow them to form internal base pairs. Their base-pairing patterns were deduced by comparing the nucleotide sequences of many species to detect conserved sequences as well as conserved base pairings. For instance, the 16S RNA of one species may have a G–C base pair, whereas another may have an A–U base pair, but the location of the base pair is the same in both molecules. Chemical modification and digestion experiments supported the structures deduced from sequence comparisons (Figure 30.15). The striking finding is that across all species, ribosomal RNAs (rRNAs) are folded into defined structures that have many short duplex regions.
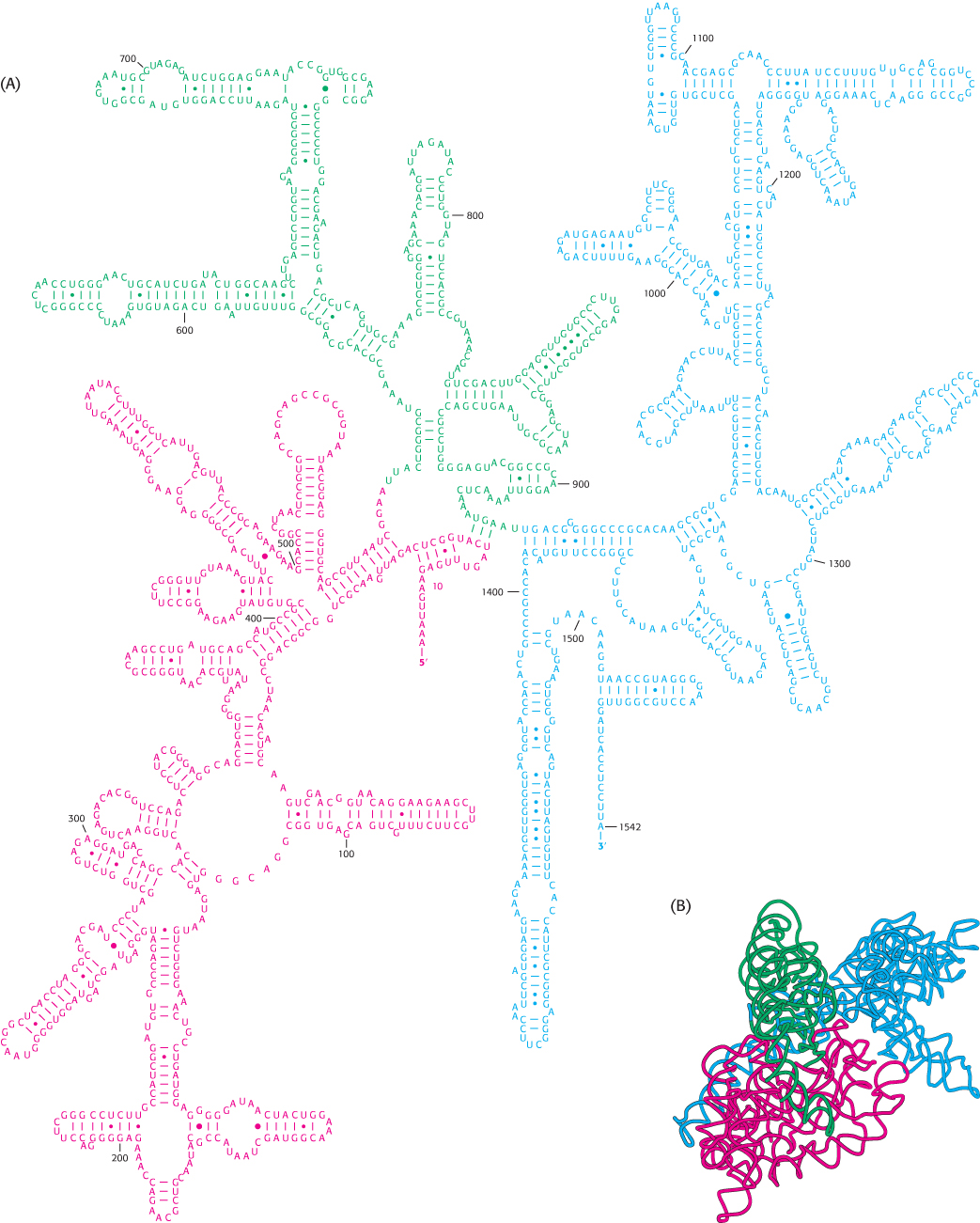
 FIGURE 30.15 Ribosomal RNA folding pattern. (A) The secondary structure of 16S ribosomal RNA deduced from sequence comparison and the results of chemical studies. (B) The tertiary structure of 16S RNA determined by x-ray crystallography.
FIGURE 30.15 Ribosomal RNA folding pattern. (A) The secondary structure of 16S ribosomal RNA deduced from sequence comparison and the results of chemical studies. (B) The tertiary structure of 16S RNA determined by x-ray crystallography.
[(A) Courtesy of Dr. Bryn Weiser and Dr. Harry Noller; (B) drawn from 1FJG.pdb.]
 For many years, ribosomal proteins were presumed to orchestrate protein synthesis and ribosomal RNAs were presumed to serve primarily as structural scaffolding. The current view is almost the reverse. The discovery of catalytic RNA (Section 29.4) made biochemists receptive to the possibility that RNA plays a much more active role in ribosomal function. The detailed structures make it clear that the key sites in the ribosome, such as those that catalyze the formation of the peptide bond and interact with mRNA and tRNA, are composed almost entirely of RNA. Contributions from the proteins are minor. The almost inescapable conclusion is that the ribosome initially consisted only of RNA and that the proteins were added later to fine-tune its functional properties. This conclusion has the pleasing consequence of dodging a “chicken and egg” question: How can complex proteins be synthesized if complex proteins are required for protein synthesis?
For many years, ribosomal proteins were presumed to orchestrate protein synthesis and ribosomal RNAs were presumed to serve primarily as structural scaffolding. The current view is almost the reverse. The discovery of catalytic RNA (Section 29.4) made biochemists receptive to the possibility that RNA plays a much more active role in ribosomal function. The detailed structures make it clear that the key sites in the ribosome, such as those that catalyze the formation of the peptide bond and interact with mRNA and tRNA, are composed almost entirely of RNA. Contributions from the proteins are minor. The almost inescapable conclusion is that the ribosome initially consisted only of RNA and that the proteins were added later to fine-tune its functional properties. This conclusion has the pleasing consequence of dodging a “chicken and egg” question: How can complex proteins be synthesized if complex proteins are required for protein synthesis?
Ribosomes have three tRNA-binding sites that bridge the 30S and 50S subunits
Three tRNA-binding sites in ribosomes are arranged to allow the formation of peptide bonds between amino acids encoded by the codons on mRNA (Figure 30.16). The mRNA fragment being translated at a given moment is bound within the 30S subunit. Each of the tRNA molecules is in contact with both the 30S subunit and the 50S subunit. At the 30S end, two of the three tRNA molecules are bound to the mRNA through anticodon–codon base pairs. These binding sites are called the A site (for aminoacyl) and the P site (for peptidyl). The third tRNA molecule is bound to an adjacent site called the E site (for exit).
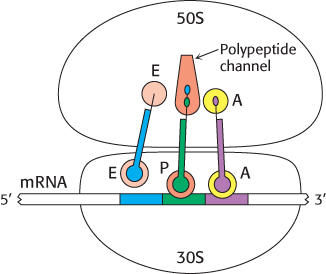
FIGURE 30.17An active ribosome. This schematic representation shows the relations among the key components of the translation machinery.
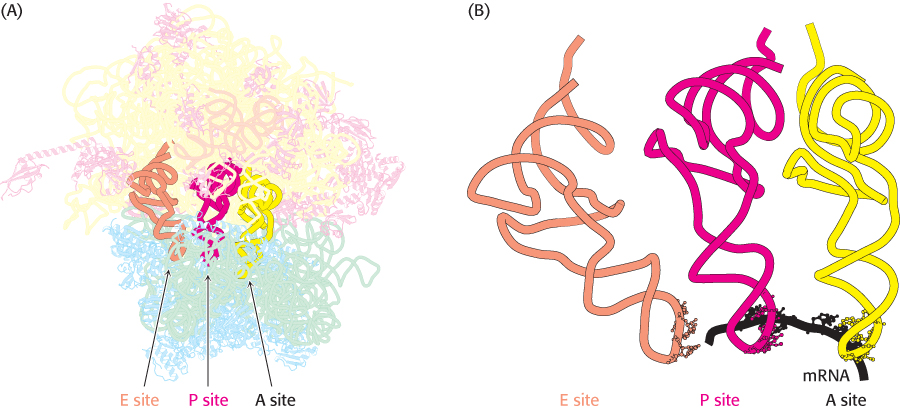
 FIGURE 30.16 Transfer RNA-binding sites. (A) Three tRNA-binding sites are present on the 70S ribosome. They are called the A (for aminoacyl), P (for peptidyl), and E (for exit) sites. Each tRNA molecule contacts both the 30S and the 50S subunit. (B) The tRNA molecules in sites A and P are base-paired with mRNA.
FIGURE 30.16 Transfer RNA-binding sites. (A) Three tRNA-binding sites are present on the 70S ribosome. They are called the A (for aminoacyl), P (for peptidyl), and E (for exit) sites. Each tRNA molecule contacts both the 30S and the 50S subunit. (B) The tRNA molecules in sites A and P are base-paired with mRNA.
[(B) Drawn from 1JGP. pdb.]
The other end of each tRNA molecule, the end without the anticodon, interacts with the 50S subunit. The acceptor stems of the tRNA molecules occupying the A site and the P site converge at a site where a peptide bond is formed. A tunnel connects this site to the back of the ribosome, through which the polypeptide chain passes during synthesis (Figure 30.17).
The start signal is usually AUG preceded by several bases that pair with 16S rRNA
How does protein synthesis start? The simplest possibility would be for the first 3 nucleotides of each mRNA to serve as the first codon; no special start signal would then be needed. However, experiments show that translation in bacteria does not begin immediately at the 5′ terminus of mRNA. Indeed, the first translated codon is nearly always more than 25 nucleotides away from the 5′ end. Furthermore, in bacteria, many mRNA molecules are polycistronic—that is, they encode two or more polypeptide chains. For example, a single mRNA molecule about 7000-nucleotides long specifies five enzymes in the biosynthetic pathway for tryptophan in E. coli. Each of these five proteins has its own start and stop signals on the mRNA. In fact, all known mRNA molecules contain signals that define the beginning and end of each encoded polypeptide chain.
A clue to the mechanism of initiation was the finding that nearly half the amino-terminal residues of proteins in E. coli are methionine. In fact, the initiating codon in mRNA is AUG (methionine) or, less frequently, GUG (valine) or, rarely UUG (leucine). What additional signals are necessary to specify a translation start site? The first step toward answering this question was the isolation of initiator regions from a number of mRNAs. This isolation was accomplished by using ribonuclease to digest mRNA–ribosome complexes (formed under conditions in which protein synthesis could begin but elongation could not take place). As expected, each initiator region usually displays an AUG codon (Figure 30.18). In addition, each initiator region contains a purine-rich sequence centered about 10 nucleotides on the 5′ side of the initiator codon.

FIGURE 30.18Initiation sites. Sequences of mRNA initiation sites for protein synthesis in some bacterial and viral mRNA molecules. Comparison of these sequences reveals some recurring features.
The role of this purine-rich region, called the Shine–Dalgarno sequence, (named after John Shine and Lynn Dalgarno, who first described the sequence), became evident when the sequence of 16S rRNA was elucidated. The 3′ end of this rRNA component of the 30S subunit contains a sequence of several bases that is complementary to the purine-rich region in the initiator sites of mRNA. Mutagenesis of the CCUCC sequence near the 3′ end of 16S rRNA to ACACA markedly interferes with the recognition of start sites in mRNA. This result and other evidence show that the initiator region of mRNA binds very near the 3′ end of the 16S rRNA. The number of base pairs linking mRNA and 16S rRNA ranges from three to nine. Thus, two kinds of interactions determine where protein synthesis starts: (1) the pairing of mRNA bases with the 3′ end of 16S rRNA and (2) the pairing of the initiator codon on mRNA with the anticodon of an initiator tRNA molecule.
Bacterial protein synthesis is initiated by formylmethionyl transfer RNA
As stated earlier, methionine is the first amino acid in many E. coli proteins. However, the methionine residue found at the amino-terminal end of E. coli proteins is usually modified. In fact, protein synthesis in bacteria starts with the modified amino acid N-formylmethionine (fMet). A special tRNA brings formylmethionine to the ribosome to initiate protein synthesis. This initiator tRNA (abbreviated as tRNAf) differs from the tRNA that inserts methionine in internal positions (abbreviated as tRNAm). The subscript “f” indicates that methionine attached to the initiator tRNA can be formylated, whereas it cannot be formylated when attached to tRNAm. Although virtually all proteins synthesized in E. coli begin with formylmethionine, in approximately one-half of the proteins, N-formylmethionine is removed when the nascent chain is 10 amino acids long.
Methionine is linked to these two kinds of tRNAs by the same aminoacyl-tRNA synthetase. A specific enzyme then formylates the amino group of the methionine molecule that is attached to tRNAf (Figure 30.19). The activated formyl donor in this reaction is N10-formyltetrahydrofolate, a folate derivative that carries activated one-carbon units (Section 24.2). Free methionine and methionyl-tRNAm are not substrates for this transformylase.
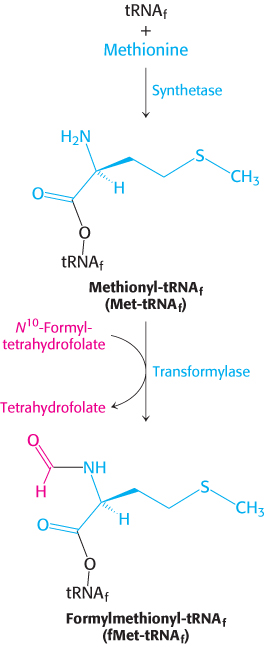
FIGURE 30.19Formylation of methionyl-tRNA. Initiator tRNA (tRNAf) is first charged with methionine, and then a formyl group is transferred to the methionyl-tRNAf from N10-formyltetrahydrofolate.
Formylmethionyl-tRNAf is placed in the P site of the ribosome in the formation of the 70S initiation complex
Messenger RNA and formylmethionyl-tRNAf must be brought to the ribosome for protein synthesis to begin. How is this task accomplished? Three protein initiation factors (IF1, IF2, and IF3) are essential. The 30S ribosomal subunit first forms a complex with IF1 and IF3 (Figure 30.20). The binding of these factors to the 30S subunit prevents it from prematurely joining the 50S subunit to form a dead-end 70S complex, devoid of mRNA and fMet-tRNAf. IF1 binds near the A site and directs the fMet-tRNAf to the P site. Initiation factor 2, a member of the G-protein family, binds GTP, and the concomitant conformational change enables IF2 to associate with fMet-tRNAf. The IF2–GTP–initiator-tRNA complex binds with mRNA (correctly positioned by the interaction of the Shine–Dalgarno sequence with the 16S rRNA) and the 30S subunit to form the 30S initiation complex. Structural changes then lead to the ejection of IF1 and IF3. IF2 stimulates the association of the 50S subunit to the complex. The GTP bound to IF2 is hydrolyzed upon arrival of the 50S subunit, releasing IF2. The result is a 70S initiation complex. The formation of the 70S initiation complex is the rate-limiting step in protein synthesis.
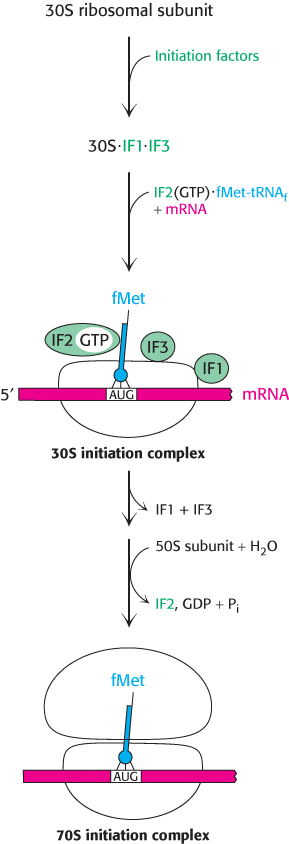
FIGURE 30.20Translation initiation in bacteria. Initiation factors aid the assembly first of the 30S initiation complex and then of the 70S initiation complex.
When the 70S initiation complex has been formed, the ribosome is ready for the elongation phase of protein synthesis. The fMet-tRNAf molecule occupies the P site on the ribosome, positioned so that its anticodon pairs with the initiating codon on mRNA. The other two sites for tRNA molecules, the A site and the E site, are empty. This interaction establishes the reading frame for the translation of the entire mRNA. After the initiator codon has been located, groups of three nonoverlapping nucleotides are defined.
Elongation factors deliver aminoacyl-tRNA to the ribosome
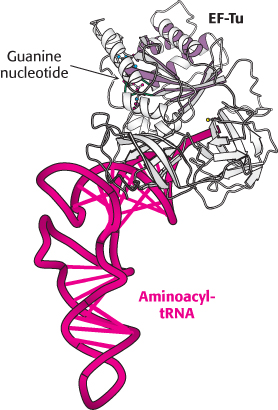
 FIGURE 30.21 Structure of elongation factor Tu. The structure of a complex between elongation factor Tu (EF-Tu) and an aminoacyl-tRNA. Notice the P-loop NTPase domain (purple shading) at the amino-terminal end of EF-Tu. This NTPase domain is similar to those in other G proteins.
FIGURE 30.21 Structure of elongation factor Tu. The structure of a complex between elongation factor Tu (EF-Tu) and an aminoacyl-tRNA. Notice the P-loop NTPase domain (purple shading) at the amino-terminal end of EF-Tu. This NTPase domain is similar to those in other G proteins.
[Drawn from 1B23. pdb.]
At this point, fMet-tRNAf occupies the P site, and the A site is vacant. The particular species inserted into the empty A site depends on the mRNA codon in the A site. However, the appropriate aminoacyl-tRNA does not simply leave the synthetase and diffuse to the A site. Rather, it is delivered to the A site in association with a 43-kDa protein called elongation factor Tu (EF-Tu), another member of the G-protein family. EF-Tu, the most abundant bacterial protein, binds aminoacyl-tRNA only in its GTP form (Figure 30.21). The binding of EF-Tu to aminoacyl-tRNA serves two functions. First, EF-Tu protects the delicate ester linkage in aminoacyl-tRNA from hydrolysis. Second, EF-Tu contributes to the accuracy of protein synthesis because GTP hydrolysis and expulsion of the EF-Tu-GDP complex from the ribosome occurs only if the pairing between the anticodon and the codon is correct. EF-Tu interacts with the 16S RNA, which monitors the accuracy of the base pairing of position 1 and 2 of the codon. Correct codon recognition induces structural changes in the 30S subunit that activate the GTPase activity of EF-Tu, releasing EF-Tu-GDP from the ribosome. These same structural changes also rotate the aminoacyl-tRNA in the A site so that the amino acid is brought into proximity with the aminoacyl-tRNA in the P site, a process called accommodation. Accommodation aligns the amino acids for peptide bond formation.
Released EF-Tu is then reset to its GTP form by a second elongation factor, elongation factor Ts. EF-Ts induces the dissociation of GDP. GTP binds to EF-Tu, and EF-Ts concomitantly departs. It is noteworthy that EF-Tu does not interact with fMet-tRNAf. Hence, this initiator tRNA is not delivered to the A site. In contrast, Met-tRNAm, like all other aminoacyl-tRNAs, does bind to EF-Tu. These findings account for the fact that internal AUG codons are not read by the initiator tRNA. Conversely, IF2 recognizes fMet-tRNAf but no other tRNA. The cycle of elongation continues until a termination codon is met.
 This GTP–GDP cycle of EF-Tu is reminiscent of those of the heterotrimeric G proteins in signal transduction (Section 14.1) and the Ras proteins in growth control (Section 14.3). This similarity is due to their shared evolutionary heritage, seen in the homology of the amino-terminal domain of EF-Tu to the P-loop NTPase domains in the other G proteins. In all these related enzymes, the change in conformation between the GTP and the GDP forms leads to a change in interaction partners. A further similarity is the requirement that an additional protein catalyzes the exchange of GTP for GDP; EF-Ts catalyzes the exchange for EF-Tu, just as an activated receptor does for a heterotrimeric G protein.
This GTP–GDP cycle of EF-Tu is reminiscent of those of the heterotrimeric G proteins in signal transduction (Section 14.1) and the Ras proteins in growth control (Section 14.3). This similarity is due to their shared evolutionary heritage, seen in the homology of the amino-terminal domain of EF-Tu to the P-loop NTPase domains in the other G proteins. In all these related enzymes, the change in conformation between the GTP and the GDP forms leads to a change in interaction partners. A further similarity is the requirement that an additional protein catalyzes the exchange of GTP for GDP; EF-Ts catalyzes the exchange for EF-Tu, just as an activated receptor does for a heterotrimeric G protein.
Peptidyl transferase catalyzes peptide-bond synthesis
With both the P site and the A site occupied by aminoacyl-tRNA, the stage is set for the formation of a peptide bond: the formylmethionine molecule linked to the initiator tRNA will be transferred to the amino group of the amino acid in the A site. The formation of the peptide bond, one of the most important reactions in life, is a thermodynamically spontaneous reaction catalyzed by a site on the 23S rRNA of the 50S subunit called the peptidyl transferase center. This catalytic center is located deep in the 50S subunit near the tunnel that allows the nascent peptide to leave the ribosome.
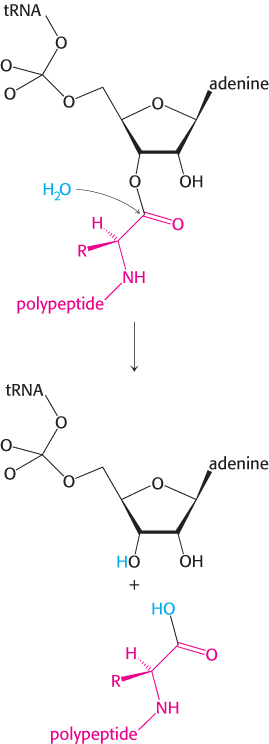
The ribosome, which enhances the rate of peptide bond synthesis by a factor of 107 over the uncatalyzed reaction (~10−4 M−1 s−1), derives much of its catalytic power from catalysis by proximity and orientation. The ribosome positions and orients the two substrates so that they are situated to take advantage of the inherent reactivity of an amine group (on the aminoacyl-tRNA in the A site) with an ester (on the initiator tRNA in the P site). The amino group of the aminoacyl-tRNA in the A site, in its unprotonated state, makes a nucleophilic attack on the ester linkage between the initiator tRNA and the formylmethionine molecule in the P site (Figure 30.22A). The nature of the transition state that follows the attack is not established and several models are plausible. One model proposes roles for the 2′OH of the adenosine of the tRNA in the P site and a molecule of water at the peptidyl transferase center (Figure 30.22B). The nucleophilic attack of the α-amino group generates an eight-membered transition state in which three protons are shuttled about in a concerted manner. The proton of the attacking amino group hydrogen bonds to the 2′ oxygen of ribose of the tRNA. The hydrogen of 2′OH in turn interacts with the oxygen of the water molecule at the center, which then donates a proton to the carbonyl oxygen. Collapse of the transition state with the formation of the peptide bond allows protonation of the 3′OH of the now empty tRNA in the P site (Figure 30.22C). The stage is now set for translocation and formation of the next peptide bond.
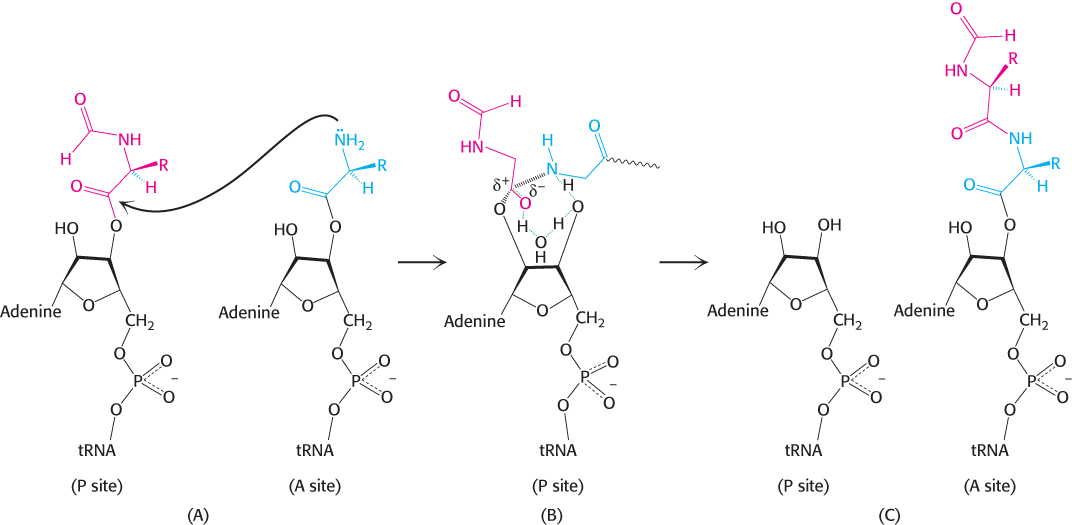
FIGURE 30.22Peptide-bond formation. (A) The amino group of the aminoacyl-tRNA attacks the carbonyl group of the ester linkage of the peptidyl-tRNA. (B) An eight-membered transition state is formed. Note: Not all atoms are shown and some bond lengths are exaggerated for clarity. (C) This transition state collapses to form the peptide bond and release the deacylated tRNA.
The formation of a peptide bond is followed by the GTP-driven translocation of tRNAs and mRNA
With the formation of the peptide bond, the peptide chain is now attached to the tRNA whose anticodon is in the A site on the 30S subunit. The two subunits rotate with respect to one another, and this structural change places the CCA end of the same tRNA and its peptide in the P site of the large subunit (Figure 30.23). Another aminoacyl-tRNA arrives and binds at the A site (1). Again, peptide bond synthesis occurs (2). However, protein synthesis cannot continue without the translocation of the mRNA and the tRNAs within the ribosome. Elongation factor G (EF-G, also called translocase) catalyzes the movement of mRNA, at the expense of GTP hydrolysis, by a distance of three nucleotides. Now, the next codon is positioned in the A site for interaction with the incoming aminoacyl-tRNA (3). The peptidyl-tRNA moves out of the A site into the P site on the 30S subunit and at the same time, the deacylated tRNA moves out of the P site into the E site and is subsequently released from the ribosome (4). The movement of the peptidyl-tRNA into the P site shifts the mRNA by one codon, exposing the next codon to be translated in the A site.
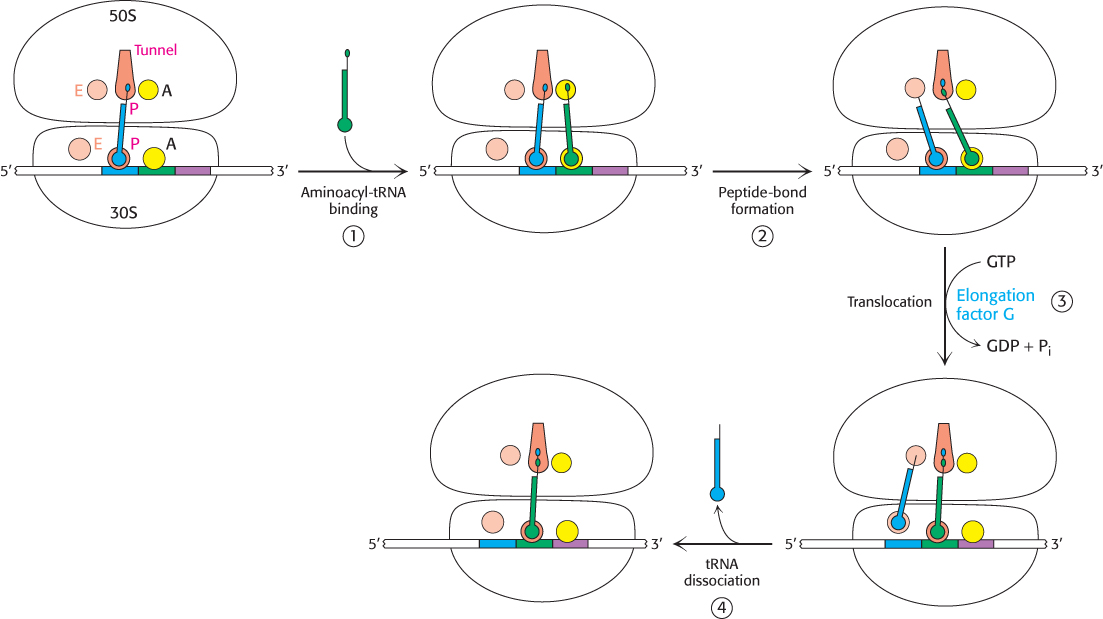
FIGURE 30.23Mechanism of protein synthesis. The cycle begins with peptidyl-tRNA in the P site. (1) An aminoacyl-tRNA binds in the A site. (2) With both sites occupied, a new peptide bond is formed. (3) The tRNAs and the mRNA are translocated through the action of elongation factor G, which moves the deacylated tRNA to the E site. (4) Once there, the tRNA is free to dissociate to complete the cycle.
The three-dimensional structure of the ribosome undergoes significant change during translocation, and evidence suggests that translocation may result from properties of the ribosome itself. However, EF-G accelerates the process. A possible mechanism for accelerating the translocation process is shown in Figure 30.24. First, EF-G in the GTP form binds to the ribosome near the A site, interacting with the 23S rRNA of the 50S subunit. The binding of EF-G to the ribosome stimulates the GTPase activity of EF-G. On GTP hydrolysis, EF-G undergoes a conformational change that displaces the peptidyl-tRNA in the A site to the P site, which carries the mRNA and the deacylated tRNA with it. The dissociation of EF-G leaves the ribosome ready to accept the next aminoacyl-tRNA into the A site.
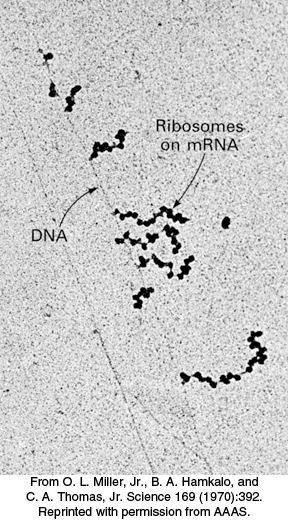
FIGURE 30.25Polysomes. Transcription of a segment of DNA from E. coli generates mRNA molecules that are immediately translated by multiple ribosomes.
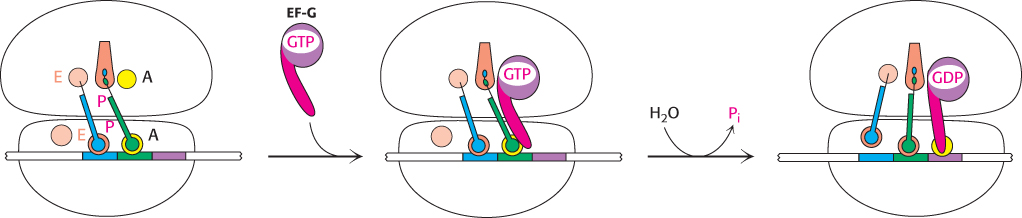
FIGURE 30.24Translocation mechanism. In the GTP form, EF-G binds to the A site on the 50S subunit. This binding stimulates GTP hydrolysis, inducing a conformational change in EF-G that forces the tRNAs and mRNA to move through the ribosome by a distance corresponding to one codon.
Note that the peptide chain remains in the P site on the 50S subunit throughout this cycle, growing into the exit tunnel. This cycle is repeated, with mRNA translation taking place in the 5′ → 3′ direction, as new aminoacyl-tRNAs move into the A site, allowing the polypeptide to be elongated until a stop signal is found.
The direction of translation has important consequences. Recall that transcription also is in the 5′ → 3′ direction (Section 29.1). If the direction of translation were opposite that of transcription, only fully synthesized mRNA could be translated. In contrast, because the directions are the same, mRNA can be translated while it is being synthesized. In bacteria, almost no time is lost between transcription and translation. The 5′ end of mRNA interacts with ribosomes very soon after it is made, well before the 3′ end of the mRNA molecule is finished. An important feature of bacterial gene expression is that translation and transcription are closely coupled in space and time.
Many ribosomes can be translating an mRNA molecule simultaneously. The group of ribosomes bound to an mRNA molecule is called a polyribosome or a polysome (Figure 30.25). Recent work shows that the ribosomes are arranged so as to protect the mRNA and to facilitate easy exchange of the substrates and products with the cytoplasm. The ribosomes in the polysome are in a helical array around the mRNA with the tRNA binding sites and peptide exit tunnel exposed to the cytoplasm.
Protein synthesis is terminated by release factors that read stop codons
The final phase of translation is termination. How does the synthesis of a polypeptide chain come to an end when a stop codon is encountered? No tRNAs with anticodons complementary to the stop codons—UAA, UGA, or UAG—exist in normal cells. Instead, these stop codons are recognized by proteins called release factors (RFs). One of these release factors, RF1, recognizes UAA or UAG. A second factor, RF2, recognizes UAA or UGA. A third factor, RF3, another GTPase, catalyzes the removal of RF1 or RF2 from the ribosome upon release of the newly synthesized protein.
RF1 and RF2 are compact proteins that, in eukaryotes, resemble a tRNA molecule. When bound to the ribosome, the proteins unfold to bridge the gap between the stop codon on the mRNA and the peptidyl transferase center on the 50S subunit (Figure 30.26). The RF interacts with the peptidyl transferase center using a loop containing a highly conserved glycine-glycine-glutamine (GGQ) sequence, with the glutamine methylated on the amide nitrogen atom of the R group. This modified glutamine (assisted by the peptidyl transferase) is crucial in promoting a water molecule’s attack on the ester linkage between the tRNA and the polypeptide chain, freeing the polypeptide chain. The detached polypeptide leaves the ribosome. Transfer RNA and messenger RNA remain briefly attached to the 70S ribosome until the entire complex is dissociated through the hydrolysis of GTP in response to the binding of EF-G and another factor, called the ribosome release factor (RRF).
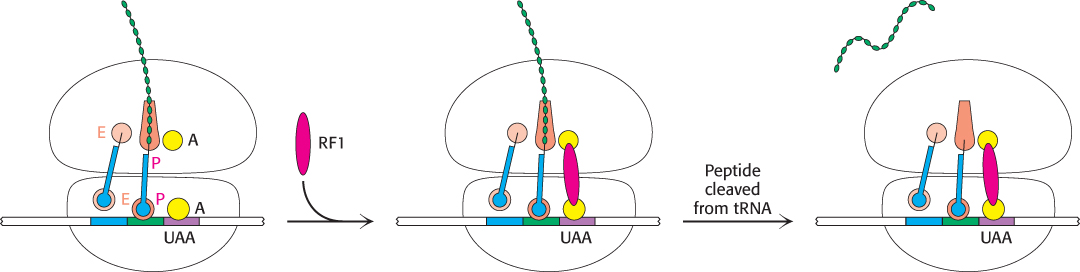
FIGURE 30.26Termination of protein synthesis. A release factor recognizes a stop codon in the A site and stimulates the release of the completed protein from the tRNA in the P site.

 FIGURE 30.14 The ribosome at high resolution. Detailed models of the ribosome based on the results of x-
FIGURE 30.14 The ribosome at high resolution. Detailed models of the ribosome based on the results of x-
 FIGURE 30.15 Ribosomal RNA folding pattern. (A) The secondary structure of 16S ribosomal RNA deduced from sequence comparison and the results of chemical studies. (B) The tertiary structure of 16S RNA determined by x-
FIGURE 30.15 Ribosomal RNA folding pattern. (A) The secondary structure of 16S ribosomal RNA deduced from sequence comparison and the results of chemical studies. (B) The tertiary structure of 16S RNA determined by x- For many years, ribosomal proteins were presumed to orchestrate protein synthesis and ribosomal RNAs were presumed to serve primarily as structural scaffolding. The current view is almost the reverse. The discovery of catalytic RNA (Section 29.4) made biochemists receptive to the possibility that RNA plays a much more active role in ribosomal function. The detailed structures make it clear that the key sites in the ribosome, such as those that catalyze the formation of the peptide bond and interact with mRNA and tRNA, are composed almost entirely of RNA. Contributions from the proteins are minor. The almost inescapable conclusion is that the ribosome initially consisted only of RNA and that the proteins were added later to fine-
For many years, ribosomal proteins were presumed to orchestrate protein synthesis and ribosomal RNAs were presumed to serve primarily as structural scaffolding. The current view is almost the reverse. The discovery of catalytic RNA (Section 29.4) made biochemists receptive to the possibility that RNA plays a much more active role in ribosomal function. The detailed structures make it clear that the key sites in the ribosome, such as those that catalyze the formation of the peptide bond and interact with mRNA and tRNA, are composed almost entirely of RNA. Contributions from the proteins are minor. The almost inescapable conclusion is that the ribosome initially consisted only of RNA and that the proteins were added later to fine-

 FIGURE 30.16 Transfer RNA-
FIGURE 30.16 Transfer RNA-



 FIGURE 30.21 Structure of elongation factor Tu. The structure of a complex between elongation factor Tu (EF-
FIGURE 30.21 Structure of elongation factor Tu. The structure of a complex between elongation factor Tu (EF- This GTP–
This GTP–