The Control of Gene Expression in Eukaryotes
CHAPTER
32
941

Many of the most important and intriguing features in modern biology and medicine, such as the pathways crucial for the development of multicellular organisms, the changes that distinguish normal cells and cancer cells, and the evolutionary changes leading to new species, entail networks of gene-
Megabase
A length of DNA consisting of 106 base pairs (if double stranded) or 106 bases (if single stranded).
1 Mb = 103 kb = 106 bases
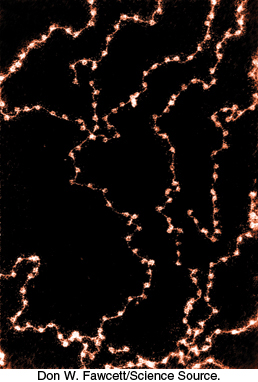
Second, whereas prokaryotic genomic DNA is relatively accessible, eukaryotic DNA is packaged into chromatin, a complex between the DNA and a special set of proteins (Figure 32.2). Although the principles for the construction of chromatin are relatively simple, the chromatin structure for a complete genome is quite complex. Importantly, in a given eukaryotic cell, some genes and their associated regulatory regions are relatively accessible for transcription and regulation, whereas other genes are tightly packaged and are thus rendered inactive. Eukaryotic gene regulation frequently requires the manipulation of chromatin structure.
942
A manifestation of this complexity is the presence of many different cell types in most eukaryotes. A liver cell, a pancreatic cell, and an embryonic stem cell contain the same DNA sequences, yet the subset of genes highly expressed in cells from the pancreas, which secretes digestive enzymes, differs markedly from the subset highly expressed in the liver, the site of lipid transport and energy transduction. Embryonic stem cells do not express any subset of genes at high levels; the most highly expressed genes are “housekeeping” genes involved in the cytoskeleton and processes such as translation (Table 32.1). The existence of stable cell types is due to differences in the epigenome, differences in chromatin structure, and covalent modifications of the DNA, not in the DNA sequence itself. Thus, different cell types share the same genome (DNA sequence) but differ in their epigenomes, the packaging and modification of this genome.
|
Rank |
Proteins expressed in pancreas |
% |
Proteins expressed in liver |
% |
Proteins expressed in stem cells |
% |
|---|---|---|---|---|---|---|
|
1 |
Procarboxypeptidase A1 |
7.6 |
Albumin |
3.5 |
Glyceraldehyde- |
0.7 |
|
2 |
Pancreatic trypsinogen 2 |
5.5 |
Apolipoprotein A- |
2.8 |
Translation elongation factor 1 α1 |
0.6 |
|
3 |
Chymotrypsinogen |
4.4 |
Apolipoprotein C- |
2.5 |
Tubulin α |
0.5 |
|
4 |
Pancreatic trypsin 1 |
3.7 |
Apolipoprotein C- |
2.1 |
Translationally controlled tumor protein |
0.5 |
|
5 |
Elastase IIIB |
2.4 |
ATPase 6/8 |
1.5 |
Cyclophilin A |
0.4 |
|
6 |
Protease E |
1.9 |
Cytochrome oxidase 3 |
1.1 |
Cofilin |
0.4 |
|
7 |
Pancreatic lipase |
1.9 |
Cytochrome oxidase 2 |
1.1 |
Nucleophosmin |
0.3 |
|
8 |
Procarboxypeptidase B |
1.7 |
α1-Antitrypsin |
1.0 |
Connexin 43 |
0.3 |
|
9 |
Pancreatic amylase |
1.7 |
Cytochrome oxidase 1 |
0.9 |
Phosphoglycerate mutase |
0.2 |
|
10 |
Bile- |
1.4 |
Apolipoprotein E |
0.9 |
Translation elongation factor 1 β2 |
0.2 |
|
Sources: Data for pancreas from V. E. Velculescu, L. Zhang, B. Vogelstein, and K. W. Kinzler, Science 270:484– |
||||||
In addition, eukaryotic genes are not generally organized into operons. Instead, genes that encode proteins for steps within a given pathway are often spread widely across the genome. This characteristic requires that other mechanisms function to regulate genes in a coordinated way.
Despite these differences, some aspects of gene regulation in eukaryotes are quite similar to those in prokaryotes. In particular, activator and repressor proteins that recognize specific DNA sequences are central to many gene-
943