Chapter Introduction
4: Protein Structure
93
MOMENT OF DISCOVERY

I’ll never forget one of our early breakthroughs in computational protein design. Our idea was to write a mathematical description of the protein structure and then optimize its thermodynamic stability by adjusting the amino acid sequence. At the time, several high-
Undaunted, we started by showing that regions of proteins could be designed using our methods. In 1996, we attempted to design a 20 amino acid polypeptide that would form a zinc finger structure, a characteristic polypeptide fold that is held together by zinc ions. After many attempts, student Bassil Dahiyat finally generated a sequence called FSD1 that was predicted to form a zinc finger fold without requiring any zinc. He synthesized this peptide in the laboratory and late that evening analyzed it by circular dichroism, a method that measures the amount of secondary structure in a protein. We had made many unsuccessful attempts at protein design by this time, so we were very familiar with the CD [circular dichroism] spectra of unfolded proteins! At about midnight, Bassil called me at home and said, “Steve, you’ve got to see this spectrum!” On my home computer over an incredibly slow Internet connection, I watched as a gorgeous spectrum with exactly the shape expected for a folded protein came up on my screen. We realized at that moment that we had achieved something many had considered impossible. When we later solved the molecular structure of the peptide using NMR spectroscopy, the peptide had exactly the structure we had predicted.
—Steve Mayo, on his discovery of the first successful method for computational protein design
94
The beauty of the DNA double helix is indisputable, but to a trained eye, protein structures are even more compelling. Proteins have wonderfully complex architectures, sculpted over time to perform their tasks to near perfection. The fact that a protein adopts a unique conformation is amazing: despite the astronomical number of ways in which even a small protein could possibly fold, it usually folds into a single shape. The instructions for the unique shape of a protein are contained entirely within the linear amino acid sequence. Exactly how the folding instructions are encoded is still not understood; it remains the holy grail of the protein-
Part of the explanation of how proteins fold lies in their reaction to an aqueous environment. Most proteins reside in the cell’s aqueous cytoplasm, yet many amino acids are hydrophobic, or water-
One might wonder why protein structures did not evolve to be more stable. In fact, thermophilic organisms—
Protein structure is commonly defined in terms of four hierarchical levels (Figure 4-1). Primary structure is essentially the sequence of amino acid residues. Secondary structure includes particularly stable hydrogen-
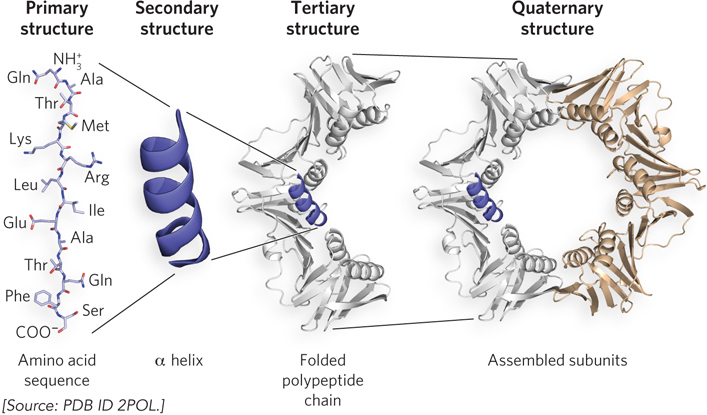
In this chapter, we explore how proteins are constructed, starting with the features of the peptide bond, which links amino acids together. Then we look at how weak forces mold protein chains into shape, and discover that, despite the bewildering array of different structures that proteins can form, all proteins contain only a few types of secondary structural elements. We will also see that there are some common ways in which these elements are stitched together to generate a diversity of folded proteins. A discussion of the two methods currently used to solve—
95